Computer Science
Peering under the “hood” of SARS-CoV-2
Microscope and protein data are incorporated into an easy-to-use-and-update tool that can model an organism’s 3D appearance.
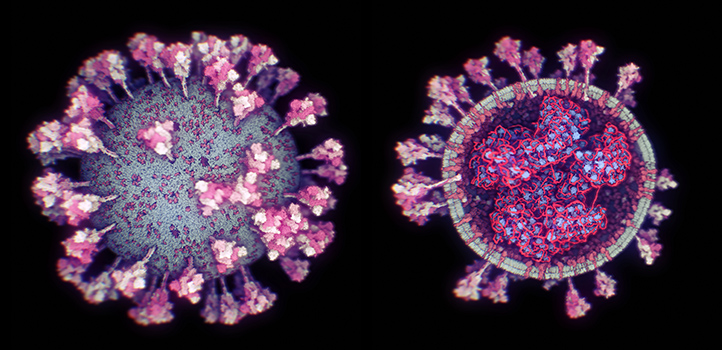
The KAUST team's 3D model demonstrates the current state-of-the-art structure of SARS-CoV-2 at the atomistic level and reveals details of the virus that were previously impossible to see. © 2020 KAUST/Nanographics
The KAUST team’s 3D model demonstrates the current state-of-the-art structure of SARS-CoV-2 at the atomistic level and reveals details of the virus that were previously impossible to see. © 2020 KAUST/Nanographics
Information from electron microscope images and protein databases has been used to develop a detailed 3D model of SARS-CoV-2, which can be readily updated as new data becomes available. The modeling tool has potential for visualizing components in other biological organisms, ranging from 10 to 100 nanometers in size.
“Our 3D model demonstrates the current state-of-the-art structure of SARS-CoV-2 at the atomistic level and reveals details of the virus that were previously impossible to see, like how we think nucleocapsid proteins form a rope-like structure inside it,” says KAUST research scientist Ondřej Strnad. “The approach we used to develop the model could steer biological research into promising new directions for fighting the spread of COVID-19, because it could help scientists rapidly incorporate newly discovered information into the model and thus provide an up-to-date structure of the virus,” he says.
The modeling system is intuitive and easy to use. It takes information from readily discernible structures in a small number of electron microscope images of an organism. For SARS-CoV-2, this involved information on the shape and size of the virus’s membrane and on the protein structures attached to it.
Scientists can then incorporate information into the model about other proteins within the cell from existing databases. This included information on SARS-CoV-2’s single RNA strand and the nucleocapsid proteins protecting it. Finally, a set of rules is created that defines how each of the components is oriented and interacts with the other components. “The system uses the provided information to predict the overall shape of the cell and generate a 3D model,” explains Strnad.
The team used their approach to develop a 3D representation of SARS-CoV-2. “Our model shows the complete viral ultrastructure as we know it to date, and not just some arbitrarily placed incomplete spike proteins on a lipid membrane,” explains KAUST computer science Ph.D. student Ngan Nguyen. “Other available models also don’t show the interior of the virus, as its details are not currently known. Scripps Research Institute, U.S., provided us with the most likely hypothesis for the structure’s interior based on current data. If this hypothesis is proven wrong, then we can easily update the model,” she explains.
The team hopes their SARS-CoV-2 model will help reveal aspects of the virus and its structure that could hasten drug discovery for treating COVID-19. They also hope scientists from different biological fields will share information and exchange rules relating to protein conformations to use the modeling system for their research.
Further improvements to the system are needed. The team plans to design a user-friendly graphical user interface to make it easy to use. They would also like to make it applicable within a virtual reality environment.
References
- Nguyen, N., Strnad, O., Klein, T., Luo, D., Alharbi, R., Wonka, P., Maritan, M., Mindek, P., Autin, L., Goodsell, D.S. & Viola, I. Modeling in the time of COVID-19: Statistical and rule-based mesoscale models. IEEE Transactions on Visualization and Computer Graphics 27, 722 -732 (2020).| article
You might also like

Computer Science
Green quantum computing takes to the skies

Computer Science
Probing the internet’s hidden middleboxes

Bioscience
AI speeds up human embryo model research
Computer Science
Improving chip design on every level

Computer Science
Sweat-sniffing sensor could make workouts smarter

Computer Science
A blindfold approach improves machine learning privacy
Computer Science
AI tool maps hidden links between diseases
Bioscience




