Computer Science
Disease researchers have a way with words
Specialized word search method sheds light on phenotypic similarity of diseases.
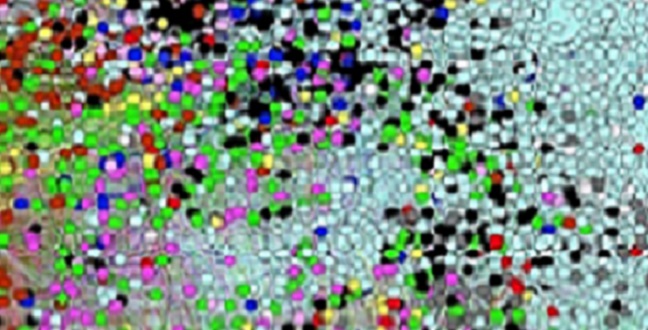
A map of the human diseasome in which nodes represent diseases and colors are based on general types of diseases. Proximity between the signs and symptoms of the diseases is used as an attraction force between the nodes so that diseases that appear close together in the map generally have similar signs and symptoms..
© 2015 KAUST
Words and phrases associated with symptoms of common diseases are used to identify complex concepts in medical texts and to point to significant patterns that help identify the genes and pathways that underlie clusters of diseases.
Researchers from KAUST worked with scientists in the United Kingdom to develop a specialized word search of scientific literature called semantic text-mining1. The work identifies shared traits in rare, common and infectious diseases for the first time at scale. It also provides, notes Robert Hoehndorf of the Computer, Electrical and Mathematical Science and Engineering Division at KAUST, “a tantalizing overview of the phenotypic structure of the human ‘diseasome.’”
Researchers routinely catalog data of signs and symptoms relating to genetically based diseases through electronic resources, such as the Online Mendelian Inheritance in Man (OMIM) and Orphanet databases. However, extending similar methods to common and infectious diseases has proved challenging due to the lack of an infrastructure providing the huge number of phenotypes associated with them.
“To take on this task, we needed very large computational capacity,” explains Hoehndorf. “Using ‘ontologies’— formal representations of the concepts and relations within a domain — we designed a method that identifies concepts referring to phenotypes of common and rare diseases within millions of published papers and abstracts, and used these concepts to establish the phenotypic similarity between a large number of common and rare diseases.”
The team’s method of grouping diseases allows for new approaches to identify the genes and pathways that underlie clusters of phenotypically similar diseases.
“If we know something about disease A, but not about disease B, finding phenotypic similarity between the two suggests they may result from a mutation or disturbance of genes or processes with a common pathway, and points to new investigations into disease B,” explains Hoehndorf.
The researchers’ work is freely available at http://aber-owl.net/aber-owl/diseasephenotypes/ (and in a visualization environment at http://aber-owl.net/aber-owl/diseasephenotypes/network/). Access to this work will allow other scientists to formulate new hypotheses about poorly understood diseases through their phenotypic similarity to others that are better characterized, or those having well-established genetic underpinnings.
“Our resource can also help to prioritize candidate genes in genome-wide and phenome-wide association studies — currently a major challenge —as well as aid the development and repurposing of drugs,” Hoehndorf explains.
References
- Hoehndorf, R., Schofield, P. N.& Gkoutos, G. V. Analysis of the human diseasome using phenotype similarity between common, genetic, and infectious diseases. Scientific Reports 5, 10888 (2015).| article
You might also like

Bioengineering
Bio-inspired network structures for next-generation AI

Computer Science
Green quantum computing takes to the skies

Computer Science
Probing the internet’s hidden middleboxes

Bioscience
AI speeds up human embryo model research
Computer Science
Improving chip design on every level

Computer Science
Sweat-sniffing sensor could make workouts smarter

Computer Science
A blindfold approach improves machine learning privacy
Computer Science



