Bioengineering
Better plant edits by enhancing DNA repair
A protein hijacked from a bacterial pathogen helps to facilitate more precise genome editing in plants.
KAUST plant scientists have developed a new genome editing system that enhances the efficiency of an error-free DNA repair pathway, which could help improve agronomic traits in multiple crops.
A new genome editing system enhances the efficiency of an error-free DNA repair pathway, which could help improve agronomic traits in multiple crops.
Genome editing involves cutting DNA at very specific locations and utilizing cells’ natural repair pathways to modify genes. Plant cells contain two repair pathways: nonhomologous end joining (NHEJ) and homology-directed repair (HDR).
“In plants, NHEJ is the dominant repair pathway at most cell stages,” says KAUST postdoc Khalid Sedeek. “But it is prone to errors. For precise genetic modifications, we need more control than provided by NHEJ. HDR is error-free but is inefficient in plants. We designed a genome editing system to enhance HDR efficiency.”

Khalid Sedeek prepares a gold particle coated with DNA for gene gun transformation.
© 2020 KAUST
The system uses Cas9 as its DNA scissor. Cas9 is fused to VirD2, a protein that comes from Agrobacterium tumefaciens, a soil bacterium that causes disease in plants by inserting part of its own DNA into the plant genome. VirD2 leads the bacterial DNA into the plant nucleus.
1
Precise engineering of plants can be achieved through binding of the VirD2 protein and the Cas9 endonuclease. Reproduced with permission under a Creative Commons License CC BY from reference 1 © 2020, Springer Nature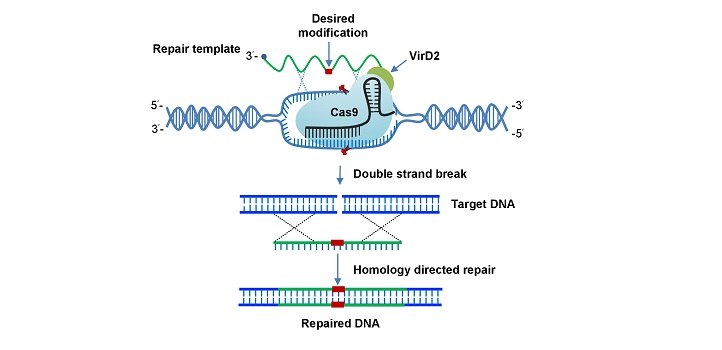
“We thought we could use VirD2 to bind to a wide range of repair templates to enhance the rate of HDR in plants by bringing the template close to the DNA break,” says KAUST research scientist Zahir Ali.
A guide RNA directs Cas9 to a specific location on the plant genome that is targeted for editing. Cas9 cuts the DNA strand, and then VirD2 holds the repair template close to the cut—the cell’s natural HDR repair pathway uses it to conduct the repair with the corresponding genetic modifications encoded in the template.
“Our data showed that the binding of the repair template to the Cas9-VirD2 system improved the rate of HDR up to 5.5 fold in plant cells compared to Cas9 alone,” says Ali.

Co-first authors Khaled Sedeek (left) and Zahir Ali evaluate the success of their genome editing system on seedlings in the KAUST Core Labs Plant Growth Chamber Facility.
© 2020 KAUST
The team tested their system in rice cells to show that the edits were precise and heritable. It successfully modified the gene that codes for the enzyme acetolactate synthase, a change that can help rice crops survive when herbicides are sprayed to kill surrounding weeds.
The system could also successfully attach a protein tag to the histone deacetylase gene in rice, which codes for an enzyme involved in plant development and reaction to stresses.
Finally, the system also successfully edited the gene that codes for the enzyme carotenoid cleavage dioxygenase 7. Precise editing of this gene results in dwarf rice plants with many grain-bearing branches.
“Our tests validated the use of this system for precise genome engineering in plants to help crop improvement,” says KAUST molecular geneticist Magdy Mahfouz, who led the study. “We expect that our Cas9-VirD2 system could be adopted across most animal and plant species.”

(r-l): Co-authors Radwa Kamel, Khaled Sedeek, Zahir Ali and Abdulrahman Alhabsi evaluate the success of their genome editing system on plants in the KAUST Core Labs Greenhouse.
© 2020 KAUST
The researchers are testing their system for simultaneous edits in several genes and for multiple edits in a single gene. They are also testing it for improving various agronomic traits in multiple crops.
References
-
Ali, Z., Shami, A., Sedeek, K., Kamel, R., Alhabsi, A., Tehseen, M., Hassan, N., Butt, H., Kababji, A., Hamdan, S.M. & Mahfouz, M.M. Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Communications Biology 3, 44 (2020).| article
You might also like
Bioengineering
Self-aware biosensors boost digital health monitoring

Bioengineering
Algae and the chocolate factory

Bioengineering
Smart patch detects allergies before symptoms strike

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities
Bioengineering
Promising patch for blood pressure monitoring

Bioengineering
Sensing stress to keep plants safe

Bioengineering
Building better biosensors from the molecule up
Bioengineering




