Bioengineering | Bioscience
Defeating the deletion makes genome editing safer and more precise
Manipulating DNA repair pathway boosts the accuracy of CRISPR technology.
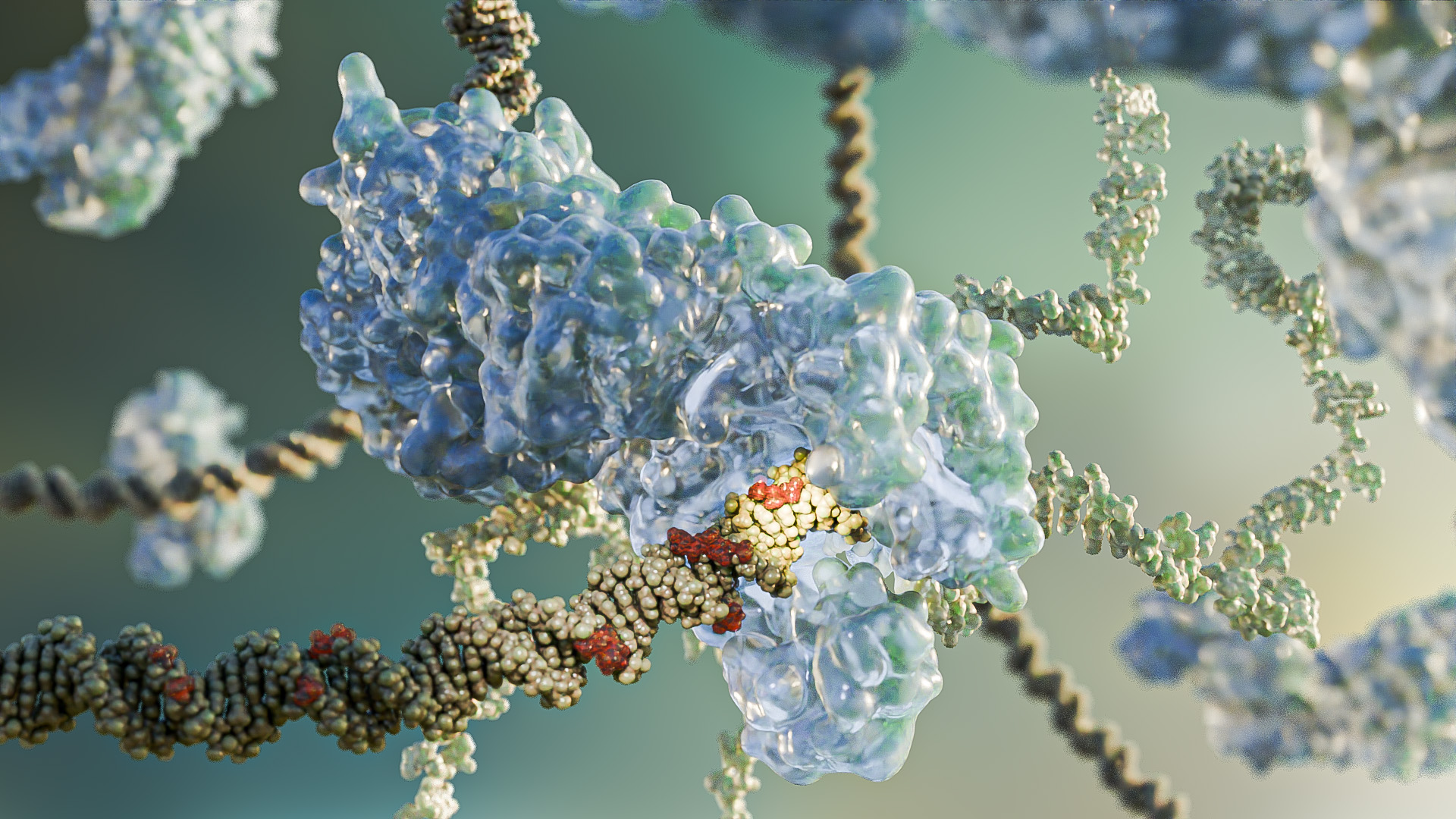
A simple and robust strategy developed by KAUST scientists could help to improve the safety and accuracy of CRISPR gene editing, a tool that is already approved for clinical use for the treatment of inherited blood disorders.
This approach tackles a critical issue with CRISPR technology: the act of slicing the genome at specific points and then rejoining it, which inherently risks damaging the DNA in a manner that might cause large-scale and unpredictable disruptions.
Hoping to mitigate this issue, a team led by Mo Li, a stem cell biologist at KAUST, investigated DNA repair pathways that lead to large genomic deletions following CRISPR editing in human stem cells.[1]
Their analysis led them to a process known as microhomology-mediated end joining (MMEJ), an error-prone mechanism that, although capable of fixing breaks in DNA, often leaves behind large deletions in its wake.
The researchers interrogated various genes implicated in this MMEJ process and found two that played central — but opposing — roles in these unwanted deletion events.
One gene, called POLQ, turned out to exacerbate the risk of large deletions following CRISPR editing. The other, called RPA, emerged as a genomic guardian with protective effects.
By manipulating these genes, either with drugs that inhibit POLQ or through genetic techniques that boost the expression of RPA, the KAUST team was then able to reduce the occurrence of detrimental large deletions without compromising the efficiency of genome editing and, in so doing, preserve the genomic integrity of edited stem cells.
“This easy-to-use approach could reduce the chances of these harmful large DNA deletions from happening,” says Baolei Yuan, a former Ph.D. student in Li’s lab and one of the architects of the study, along with Chongwei Bi and Yeteng Tian from Li’s lab.
Moreover, these same interventions were found to enhance the efficiency of homology-directed repair, a mechanism known for its ability to enable accurate genome editing without adding unintended mutations.
This was evident in experiments involving stem cells that carried mutations in two genes linked to sickle cell disease and Wiskott-Aldrich Syndrome, both inherited blood disorders. By modulating POLQ or RPA, the researchers achieved highly precise and reliable gene editing in these cells.
The findings mark a significant step forward in refining CRISPR technology, asserts Li. “It’s really exciting because it means we’re getting closer to safer and more effective treatments for genetic diseases,” he says.
With a provisional patent application filed for this innovative strategy, the team continues to explore the mechanisms behind a wider array of undesirable mutations and to hone its techniques for making CRISPR safer and more efficient.
“Achieving both high efficiency and safety remains a challenge that requires further development,” Li says, “and our laboratory remains at the forefront, seeking out novel solutions.”
Reference
- Yuan, B., Bi, C., Tian, Y., Wang, J., Jin, Y., Alsayegh, K., Tehseen, M., Yi, G., Zhou, X., Shao, Y., Vargas Romero F., Fischle, W., Izpisua Belmonte, J.C., Hamdan, S., Huang, Y. & Li, M. Modulation of the microhomology-mediated end joining pathway suppresses large deletions and enhances homology-directed repair following CRISPR-Cas9-induced DNA breaks. BMC Biology (2024).| article
You might also like
Bioengineering
Self-aware biosensors boost digital health monitoring

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioengineering
Algae and the chocolate factory

Bioengineering
Smart patch detects allergies before symptoms strike

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience




