Bioengineering | Bioscience
Pioneering technique transforms genetic disorder diagnoses
An efficient long-read sequencing technique that searches for complex genomic variants provides accurate molecular diagnosis for genetic disorders within hours.
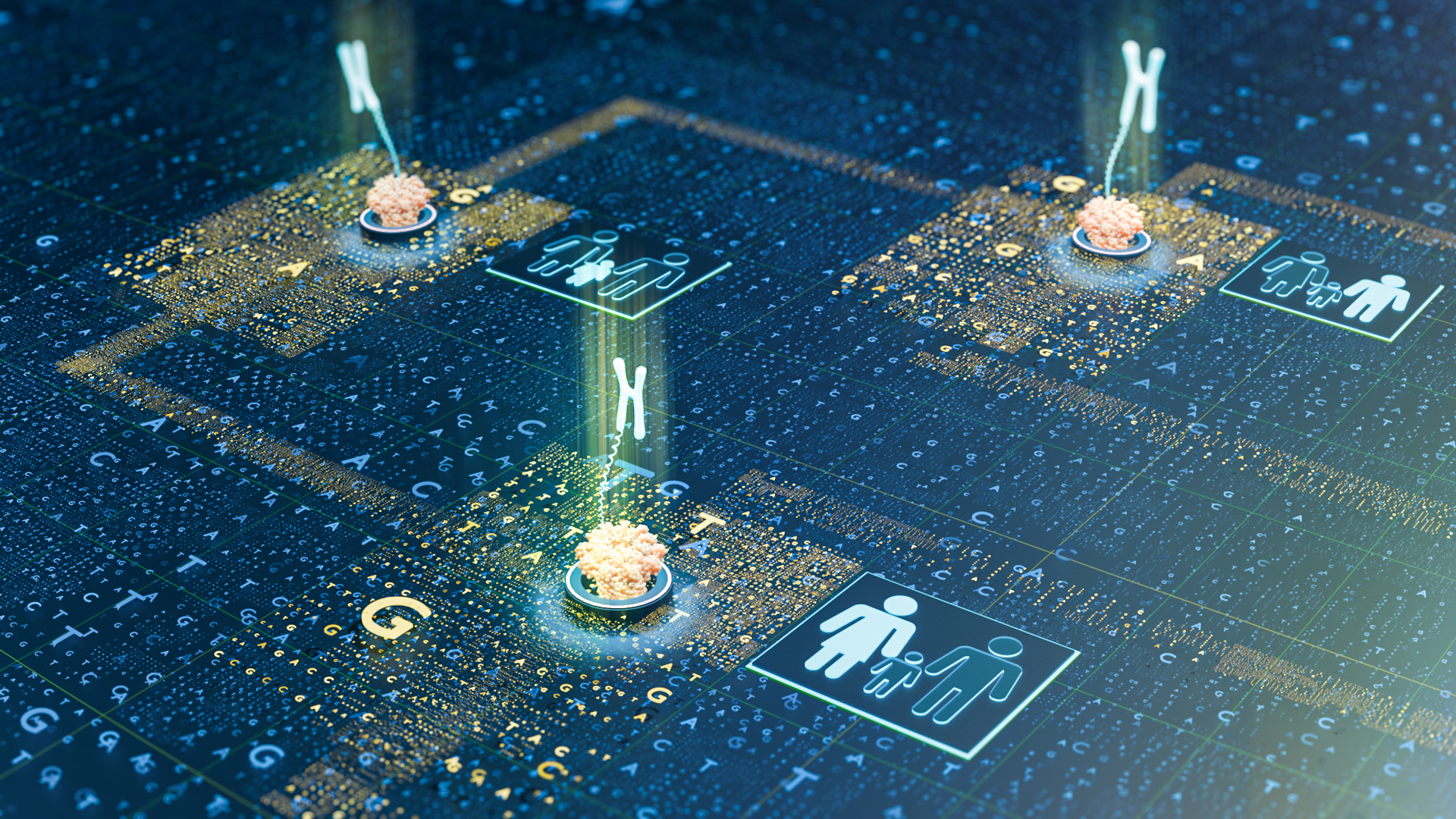
Despite rapid advances in genetic testing in recent decades, more than half of people worldwide with suspected Mendelian genetic disorders do not have an accurate molecular diagnosis. Others endure more than six years of tests before a diagnosis is given. Now, KAUST researchers and scientists across Saudi Arabia have developed NanoRanger, an accurate and rapid method for genetically diagnosing such diseases in a few hours[1].
“Precise, efficient genomic diagnosis is urgently needed to improve patient outcomes and facilitate carrier screening,” says Yingzi Zhang, a Ph.D. candidate at KAUST, supervised by Mo Li. “This study aligns with Saudi Arabia’s Vision 2030 — advancing healthcare through innovation to improve quality of life for all citizens.”
Mendelian disorders — including nervous system and intellectual developmental conditions — are caused by either an alteration in one particular gene or an abnormal rearrangement in one segment of the genome. Each disease has a specific “breakpoint”— the genomic location of a structural variant where DNA is deleted, rearranged or inverted.
While these variants may be identified using traditional screening techniques, the sheer complexity of the rearrangements means they are often missed. Mendelian diseases are inheritable, particularly if both parents are carriers of the same faulty segment. Such diseases are more prevalent in regions where it is common for marriage between related individuals (consanguinity).
“NanoRanger uses simple molecular biology strategies to ‘fish out’ genomic regions that are suspected of harboring complex mutations, deletions or rearrangements,” says Li.
The technique is cost-effective and requires only a tiny amount of DNA from a patient or suspected carrier. NanoRanger takes a sample of genomic DNA and uses molecular scissors called restriction enzymes to fragment the DNA into pieces with the same end sequences. These pieces are then self-joined into circles and amplified, which makes it easier to target and sequence the genomic regions of interest using Oxford Nanopore Tecnologies’ long-read sequencing technology.
“Using our custom-developed data analysis tool, NanoRanger accurately maps breakpoints at single base-pair resolution, providing a detailed picture that helps diagnose the genetic disorder,” says Zhang. “Diagnosis can be as fast as 12 minutes after initial sequencing, which is a game-changer.”
In trials done in collaboration with a group of Saudi clinicians led by Fowzan Alkuraya at King Faisal Specialist Hospital & Research Center, NanoRanger successfully identified precise breakpoints in 13 familial cases of genomic disorders that were missed by conventional genetic tests. Using these breakpoints, the researchers then screened the carrier status of related family members and 1,000 healthy Saudi individuals.
The testing method prompted one Saudi couple in the trial to opt for in vitro fertilization after they were both found to carry the genomic deletion for an inherited Mendelian disease.
“We have filed for a patent, and plan to integrate NanoRanger into standard diagnostic routines to provide a comprehensive toolkit for clinical settings, both here in Saudi Arabia and across the world,” concludes Li.
Reference
- Zhang, Y., Bi, C., Nadeef, S., Maddirevula, S., Alqahtani, M., Alkuraya, F.S. & Li, M. NanoRanger enables rapid single base-pair resolution of genomic disorders. Med (2024).| article
You might also like
Bioengineering
Self-aware biosensors boost digital health monitoring

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioengineering
Algae and the chocolate factory

Bioengineering
Smart patch detects allergies before symptoms strike

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience




