Bioengineering
Pathogens hitch a lift on host’s metabolism
A new computational framework shows how pathogens interact with host cells at the metabolic level.
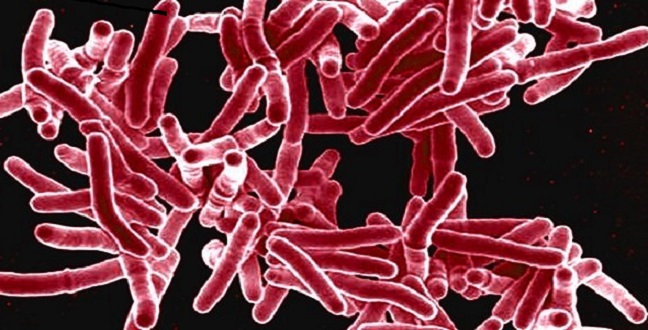
When a pathogen infects a host cell, it starts to take over — or ‘hijack’— some of the cell’s processes. The network of chemical reactions that comprise the host’s metabolism is forced to make essential products for pathogen survival, growth or reproduction. Deciphering this interplay is complex, but KAUST researchers have developed a way to explore a pathogen’s route to overtaking their host’s metabolism1.
The team, led by Dimitrios Kleftogiannis, designed a series of programs known as ‘Hi-Jack’, which identify potential connectivity points between the metabolic pathways of host and pathogen. This approach has only recently become possible as new metabolic data is delivered. Kleftogiannis says data availability is vital for such analysis. “We are lucky that, with recent advances in biotechnology and high-throughput methods, metabolic network data are growing rapidly.”
Hi-Jack compares the metabolic networks of host and pathogen to identify host pathways which produce molecules required by the pathogen. The potential of these candidate pathways as hijacking targets are then ranked.
The team tested Hi-Jack on the interaction between the tuberculosis bacterium, Mycobacterium tuberculosis, and humans. It compared 287 human and 115 bacterial metabolic pathways, representing a total of nearly 1000 reactions.
Thirty-six human pathways were identified as potential hijacking targets, each providing multiple compounds useful to M. tuberculosis. These pathways all scored highly against a set of validation principles, such as production of proteins essential for bacterial growth and presence in host tissues that are colonized by the pathogen. This indicates that Hi-Jack correctly identified potential hijacking targets.
The strongest candidate pathways for hijacking were those for carbohydrate, amino acid, and lipid metabolism. These would provide M. tuberculosis with energy and raw materials for growth, and are strongly associated with the lungs and lymph nodes: key sites of tuberculosis infection.
Interestingly, more than 90 percent of the pathways were also targets for existing tuberculosis drugs. In future, Hi-Jack may be able to predict potential drug targets, and the KAUST team is investigating the possibility.
By improving understanding of the interplay between host and pathogen, Hi-Jack could support new strategies for prevention and treatment of debilitating and increasingly drug-resistant diseases such as tuberculosis, leprosy, flu and even HIV. Kleftogiannis points out that the only constraint to development is data availability, but a rapidly growing metabolic data network is removing this obstacle, indicating Hi-Jack will find a much wider application in the future.
References
- Kleftogiannis, D., Wong, L., Archer, J.A.C. & Kalnis, P. Hi-Jack: a novel computational framework for pathway-based inference of host-pathogen interactions. Bioinformatics 31, 2332-2339 (2015).| article
You might also like
Bioengineering
Self-aware biosensors boost digital health monitoring

Bioengineering
Algae and the chocolate factory

Bioengineering
Smart patch detects allergies before symptoms strike

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities
Bioengineering
Promising patch for blood pressure monitoring

Bioengineering
Sensing stress to keep plants safe

Bioengineering
Building better biosensors from the molecule up
Bioengineering



