Bioengineering
Decoding noncoding sequences
Evolution has left alone the protective functions of noncoding sequences of the human genetic blueprint.
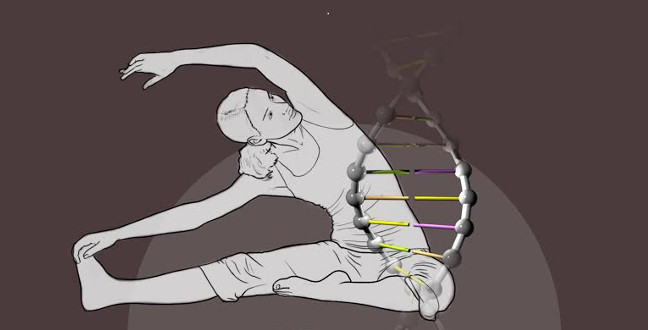
Human and insect genomes have many noncoding sequences in common, most of which are unchanged since evolution began. As their longevity indicate essential roles, KAUST researchers analyzed these noncoding sequences to better understand what they do and how they manage to withstand evolutionary change.
Timothy Ravasi and colleagues from KAUST’s Division of Biological and Environmental Science and Engineering and Division of Computer, Electrical, and Mathematical Science and Engineering carried out an integrative analysis of the genome of the classic insect model, fruit fly Drosophila melanogaster1. “Our findings suggest that genome architectures are more complex than we anticipated,” says Ravasi. “By identifying repeated and very conserved elements we are able to shed new light on the mechanisms of genomes evolution and regulation.”
The genome, as an organism’s complete set of genetic instructions, contains all the coding sequences needed to build that organism from scratch. It also contains noncoding sequences, whose mutations are implicated in diseases, including neurodevelopmental disorders and cancer. Understanding these noncoding sequences helps scientists to pinpoint productive areas for new therapeutic inquiry.
Ravasi’s team studied the highly conserved noncoding sequences throughout the development of D. melanogaster, focusing on the sequences’ genomic properties, transcriptional activities and epigenetic regulations. Their results showed that histone modifications — for example, lysine methylation and acetylation — of highly conserved noncoding sequences can modulate the transcription of nearby genes, particularly those genes that regulate development.
They identified 1,456 highly conserved noncoding sequences. These sequences were shown to have distinct features, such as higher guanine–cytosine content, when compared to other noncoding sequences. In addition, they showed that highly conserved noncoding sequences reside in compact DNA molecules — or chromatin structures — and exhibit fewer histone modifications during development than during embryonic or larval stages. As a result, the transcriptional activities of adjacent genes are highest during development. Considered together, the findings suggest that highly conserved noncoding genes and their histone modifications are involved in the transcriptional regulation of adjacent genes.
The researchers also examined the effects of highly conserved noncoding sequences on the timing of DNA replication — a finely orchestrated process that ensures genetic instructions are correctly transferred during cell division. Although 10 percent of highly conserved noncoding sequences were shown to fire early in replication, most fired relatively late.
“Our finding suggests that highly conserved noncoding sequences are under selective pressures, which helps explains how they manage to remain highly conserved,” says Ravasi.
References
- Seridi, L., Ryu, T. & Ravasi, T. Dynamic epigenetic control of highly conserved noncoding elements. PLOS ONE 9, e109326 (2014).| article
You might also like
Bioengineering
Self-aware biosensors boost digital health monitoring

Bioengineering
Algae and the chocolate factory

Bioengineering
Smart patch detects allergies before symptoms strike

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities
Bioengineering
Promising patch for blood pressure monitoring

Bioengineering
Sensing stress to keep plants safe

Bioengineering
Building better biosensors from the molecule up
Bioengineering



