Marine Science
Giant bacteria make algae easy to stomach
Symbiotic giant bacteria enable Red Sea surgeonfish to specialize their diets.

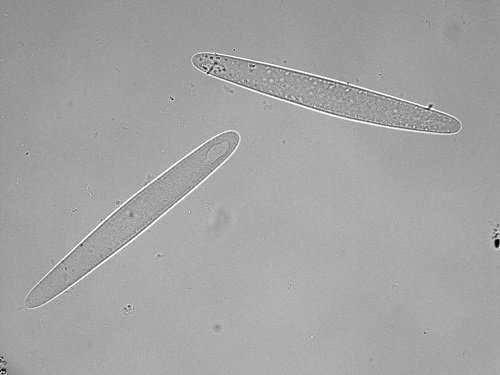
Epulopiscium bacteria are helping to explain the diversity of surgeonfish and the specialization of their diet.
© 2017 Sou Miyake

Surgeonfish Naso elegans are reported to feed on macroscopic brown algae.
© 2017 Till Rothig and Anna Roik

Acanthurus sohal are reported to feed on either turfing or filamentous red and green algae.
© 2017 Till Rothig and Anna Roik
Red Sea surgeonfish use metabolically diverse giant bacteria to digest different types of algae, according to new research. Not only do these findings explain the basis of surgeonfish diversity, but they may also provide a valuable genetic resource for biofuel research.
An international team led by KAUST researchers used high-throughput sequencing techniques to study symbiotic microbe communities in the intestines of marine-algae-feeding Red Sea surgeonfish. By analyzing the genomes, they discovered that the communities are dominated by a single group of giant bacteria known as Epulopiscium, and that they lack the diversity found in the microbiomes of terrestrial herbivores.
“The degradation of plant biomass in terrestrial vertebrates usually requires cocktails of enzymes originating from gut microorganisms, each of which has the capacity to break down different constituents,” explains KAUST research scientist David Ngugi, who led the study. Algae lack many of the complex cell wall constituents and polysaccharides found in land plants, such as lignin and cellulose, and so a simpler microbial community is likely sufficient to digest them.
Nevertheless, analyzing gene expression revealed major differences between the Epulopiscium in surgeonfish specialized by feeding on red or brown algae. “Depending on the algae that the host is feeding on, the Epulopiscium have corresponding enzymes to break down those polysaccharides,” says Ngugi. “So much so that you probably cannot take an Epulopiscium from a red-algae eating host and transplant it to brown-algae eating host because they don’t have the metabolic capacity to degrade what the other host is eating.”
This specialization helps explain the diversity of reef surgeonfish because they divide the environment into different dietary niches. Based on their analyses, the researchers suggest dividing the symbiotic Epulopiscium into three genera.
The team also tracked gene expression in Epulopiscium throughout a day and found that it matched the host’s lifestyle, with genes related to digestion active during the morning when the host was feeding. “That was really exciting,” says Ngugi, because it clearly demonstrated the giant bacteria’s role in the gut.
The ability to ferment algae will make Epulopiscium a valuable genetic resource for the development of algal-based biofuels. The findings also highlight the link between food, fish and microbes underpinning the diversity of reef communities. “The kind of symbiosis that has developed in order to utilize the specific food resources in the reef has occurred over evolutionary time scales,” says Ngugi. “Our data suggest that it’s not something that can be acquired or re-established in a short time.”
References
-
Ngugi, D.K., Miyake, S., Cahill, M., Vinu, M., Hackmann, T., Blom, J., Tietbohl, M., Berumen, M.L. & Stingl, U. Genomic diversification of giant enteric symbionts reflects host dietary lifestyles. Proceedings of the National Academy of Sciences 114, E7592–E7601 (2017).| article
You might also like

Marine Science
Measuring spatial differences in reef-building corals to guide future management

Marine Science
Potential gains from replenishing reef fish stocks revealed

Marine Science
A place to trial hope for global reef restoration

Marine Science
Reef-building coral shows signs of enhanced heat tolerance

Marine Science
Plastic-munching bacteria found across the seven seas

Marine Science
AI reveals the universal beauty of coral reef growth

Marine Science
Tiny crabs glow to stay hidden

Marine Science



