Computer Science
Sharing expert experimental knowledge to expedite design
A repository of metabolic information provides a quick reference tool for designing useful synthetic biological systems.
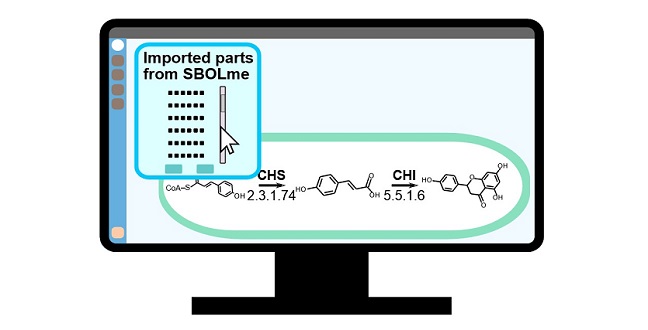
An SBOL-compliant natural product biosynthesis design tool
© 2017 KAUST
A reference tool specific to metabolic engineering that optimizes processes to make cells produce useful substances gives researchers a common language and will facilitate novel designs.
When building synthetic biological systems, the number of available biological components and interactions is mindboggling. To make them accessible, a global community of researchers developed the Synthetic Biology Open Language (SBOL), an online reference tool that allows experimental and computational biologists to quickly exchange and reuse designs1.
Now, Hiroyuki Kuwahara and Xin Gao of the KAUST Computational Bioscience Research Center (CBRC) collaborated with co-workers in China and the US1 to take this a step further by developing an SBOL repository of parts specific to metabolic engineering.
“So far, support for SBOL has been lacking in metabolic engineering,” said Kuwahara. “But, since many applications of synthetic biology focus on biosynthesis of high-value natural products, such as biofuels, cosmetics, perfumes and drugs, we thought that a repository of SBOL parts for metabolic engineering would be useful.”
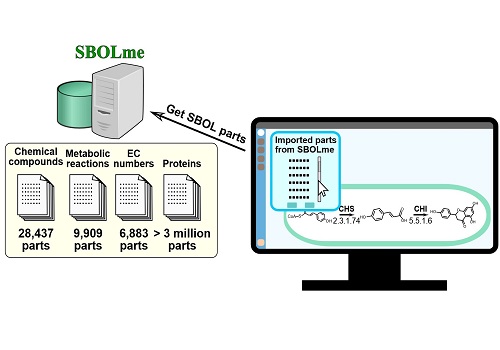
Kuwahara, Gao and co-workers drew on several existing databases to compile the new, easily searchable repository of metabolic parts, which they named SBOLme, because it is entirely compliant with SBOL.
SBOLme contains several thousand chemical compounds, enzyme classes, metabolic proteins and metabolic reactions from almost 4000 different organisms, and all the parts are annotated with information about their common interactions and thermodynamics. This will make it possible for researchers to design new ways of carrying out useful reactions, as Kuwahara explained, “Suppose you want to design a pathway to produce 1,3-propanediol, a commodity chemical mainly used to make polyester fiber from glycerol that is readily available and inexpensive. This would involve introducing two enzymatic reactions into a microbe that could perform the job. SBOLme has all the metabolic parts you need to specify this pathway within the microbe, and it allows you to exchange and discuss the design with colleagues.”
“Finally, when you publish your paper, you would include your design in the SBOL format so that readers can find it.”
Kuwahara and Gao say that the talented computer scientists at KAUST and the CBRC’s cluster computing facilities played a vital role in the success of their project.
They hope to continue developing computational methods that help facilitate the design of metabolic engineering. Ultimately, they plan on making the industrial-scale use of microbes for producing useful chemicals accessible.
The potential applications of metabolic engineering include the production of new drugs and the storage of information on strands of DNA.
References
- Kuwahara, H., Cui, X., Umarov, R., Grünberg, R., Myers, C.J. & Gao, X. SBOLme: a repository of SBOL parts for metabolic engineering. ACS Synthetic Biology 6, 732-736 (2017).| article
You might also like

Bioengineering
Bio-inspired network structures for next-generation AI

Computer Science
Green quantum computing takes to the skies

Computer Science
Probing the internet’s hidden middleboxes

Bioscience
AI speeds up human embryo model research
Computer Science
Improving chip design on every level

Computer Science
Sweat-sniffing sensor could make workouts smarter

Computer Science
A blindfold approach improves machine learning privacy
Computer Science



