Bioengineering | Bioscience
Automated COVID-19 diagnosis, with wider potential
An open-source, automated and cost-effective platform for COVID-19 diagnosis and analysis of genetic variants can be built from readily available materials, with big possibilities on the horizon.
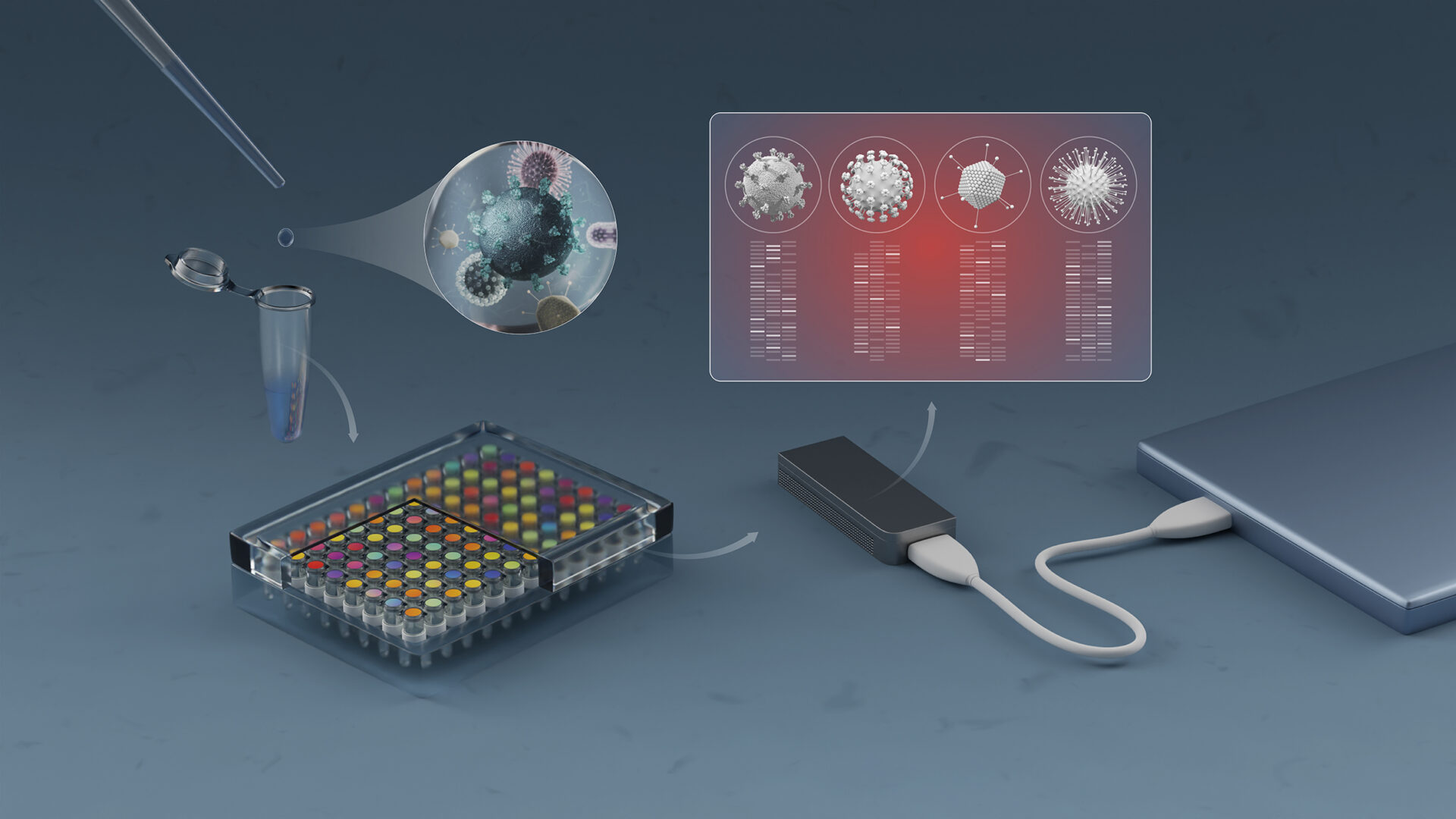
As the COVID-19 pandemic swept the globe in early 2020, a group of researchers at KAUST repurposed readily available equipment and chemicals to produce a homegrown robotic system to extract and sequence the viral RNA.[1] The system could also be used to track the emergence of new virus variants, be applied to other viral threats and perhaps eventually diagnose genetic disorders.
“With global supply chains broken or unreliable, we wanted to see whether we could utilize our research lab facilities for an emergency response and also produce all the necessary materials ourselves rather than relying on commercial vendors from overseas,” says research scientist Raik Grünberg. “We are now much more ready for the next pandemic.”
Several KAUST teams worked together on the project under the umbrella of the KAUST Smart Health Initiative. Mo Li and Gerardo Ramos-Mandujano developed magnetic beads for extracting viral RNA. Grünberg and colleagues repurposed an existing liquid-handling robot to automate RNA extraction. Samir Hamdan’s lab purified and tested the enzymes required to detect and quantify the viral RNA.
As the importance of new SARS-CoV-2 virus variants became apparent, the team refocused their efforts from merely testing for the virus to also tracking the mutations driving viral evolution. They integrated KAUST’s own sequencing technology, called NIRVANA (Nanopore Sequencing of Isothermal Rapid Viral Amplification for Near Real-time Analysis). NIRVANA was previously developed by Li and colleagues and can be performed on a hand-held device that requires very little lab infrastructure.
Bioscientist Mo Li points out: “It’s great to know that KAUST can quickly assemble a rapid, high-throughput and comprehensive diagnosis platform completely based on Saudi-developed technologies.”
Li’s colleague Yingzi Zhang adds: “The final result is a portable and affordable system that can be produced from parts and materials that are likely to be readily available in many labs, including those in regions of the world with very limited research budgets.”
All the information needed to build the system is freely available after open-source publication, removing any commercial barriers to its use and fostering further improvements by others.
Zhang says that the team is, however, considering some prospects for commercialization, “building on the protocol that we developed for magnetic-nanoparticle-aided viral RNA isolation from contagious samples.” This protocol has received approval from the Saudi Food and Drug Authority (SFDA).
In addition to detecting disease-causing pathogens, the team now hopes to extend the application of the technology into diagnosing genetic disorders, opening a whole new field of use.
Reference
- Ramos‑Mandujano, G., Grünberg, R., Zhang, Y., Bi, C., Guzmán‑Vega, F. J., Shuaib, M., Gorchakov, R. V., Xu, J., Tehseen, M., Takahashi, M., Takahashi, E., Dada, A., Ahmad, A. N., Hamdan, S. M., Pain, A., Arold, S. T. & Li, M. An open‑source, automated, and cost‑effective platform for COVID‑19 diagnosis and rapid portable genomic surveillance using nanopore sequencing. Scientific Reports 13, 20349 (2023).| article.
You might also like
Bioengineering
Self-aware biosensors boost digital health monitoring

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioengineering
Algae and the chocolate factory

Bioengineering
Smart patch detects allergies before symptoms strike

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience




