Bioscience
DNA replication under the microscope
The three-dimensional structure of a protein complex that is key for replicating damaged DNA offers insights into its regulation.
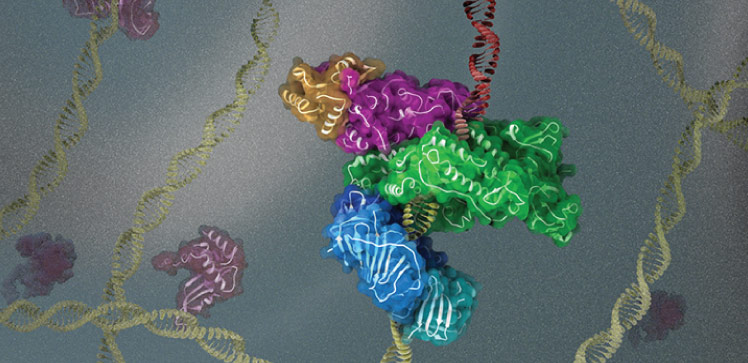
Cryogenic electron microscopy (cryo-EM) has enabled researchers to study how the DNA replication machinery assembles at sites where DNA is damaged.
Cellular DNA is continuously exposed to both endogenous and exogenous DNA-damaging agents, such as reactive oxygen species and UV radiation. To reduce the biological consequences of DNA damage, all living organisms have evolved mechanisms to tolerate and repair DNA damage to try to ensure that genetic information is accurately inherited. One such mechanism, called translesion synthesis (TLS), allows DNA replication to proceed through unrepaired DNA lesions.
TLS involves highly accurate DNA synthesizing enzymes (replicative DNA polymerases) being temporarily replaced with specialized, low-fidelity TLS polymerases that can ensure cell survival at the expense of introducing mutations. The mutagenic and translesion synthetic activity of TLS polymerases can result in normal cells becoming cancerous or cancer cells becoming drug resistant.
The Y-family TLS polymerase Pol K is able to perform DNA synthesis across several damaged bases and is recruited to DNA lesions by proliferating cell nuclear antigen (PCNA). Previous studies have shown that PCNA is regulated by ubiquitination. “The addition of a single ubiquitin molecule at lysine residue 164 (K164) of PCNA facilitates the recruitment and retention of TLS polymerases to damage sites, but the structural basis of the interaction between these polymerases and ubiquitinated PCNA is poorly understood,” says structural biologist Alfredo De Biasio from KAUST.
Since 2018, De Biasio’s group has been collaborating with the laboratory led by Samir Hamdan, an expert in single-molecule analysis of human DNA replication. They have been using cryo-EM to investigate the three-dimensional structure and function of key protein complexes involved in DNA replication and repair.
Their latest study describes cryo-EM reconstructions of full-length human Pol K bound to DNA, an incoming nucleotide, and unmodified PCNA or mono-ubiquitylated PCNA, at near-atomic resolution. They found that in the absence of DNA, the structure of Pol K bound to PCNA is highly flexible, suggesting that binding to DNA is required to form a rigid and active complex.
Muhammad Tehseen, a senior researcher in Hamdan’s group and the co-lead author of the study, has performed key functional studies elucidating how PCNA ubiquitination modulates the activity of Pol K.
“Our data provide a structural framework to explain how PCNA recruits a Y-family TLS polymerase to sites of DNA damage,” Tehseen explains. Because of the high degree of domain conservation between Y-family polymerases, some of the structural features observed in the Pol K-DNA-PCNA complex are likely to apply to other TLS polymerase complexes.
“By understanding the interactions between the proteins forming these complexes and how they are regulated, we can identify ways to reduce or increase their function for medical applications,” he concludes.
References
- Lancey, C., Tehseen, M., Bakshi, S., Percival, M., Takahashi, M., Sobhy, M.A., Raducanu, V.S., Blair, K., Muskett, F.W., Ragan, T.J., Crehuet, R., Hamdan, S.M. & De Biasio, A. Cryo-EM structure of Pol k bound to DNA and mono-ubiquitylated PCNA. Nature Communications 12, 6095 2021.| article
You might also like

Bioengineering
Bio-inspired network structures for next-generation AI

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience




