Bioscience
Perfect conditions for precision nanostructures
Protein-folded DNA nanostructures offer a new building material for biotechnology.
KAUST bioscientists have characterized the structure of histone-DNA nanostructures.
©2019 KAUST
By using proteins that naturally bind and arrange DNA inside cells, a KAUST-led team has devised a plug-and-play strategy for building stable, custom-designed nanostructures.
The versatile yet straightforward method for designing hybrid DNA-protein assemblages now provides engineers with a nanoscale platform for solving problems in science. “The DNA-protein nanotechnology has potential applications in many fields, including medicine, biotechnology and analytical chemistry,” says KAUST’s Professor Satoshi Habuchi, who led the study.
The idea of using DNA as a kind of molecular origami dates back to the 1980s, but it was only two years ago that scientists succeeded at incorporating proteins into nanostructures. As such a nascent technology, Habuchi realized the scope for improvement, which he identified, “Demanded the finding of new building blocks for the construction of DNA-protein self-assembled nanostructures.

Maged Serag (left) and Hubert Piwoński discuss the results of their single-molecule fluorescence microscopy.
© 2019 KAUST
The building block that Habuchi and his team chose to incorporate into their structures is called a histone, a type of protein that normally acts like a spool to wind and compact DNA inside the cell. Under the right artificial conditions, histones and single-stranded DNA will also spontaneously self-assemble into individual nanoparticles and cross-linked complexes.
Using electron microscopes at the University’s Imaging and Characterization Core Lab and other cutting-edge equipment in Habuchi’s lab, the researchers characterized the structure of these histone-DNA nanostructures. They were able to detail how they form with precise geometry if given the right combination of temperature, incubation time and chemical environment.
The only variable that seemed to change shape was the length of the DNA.

These images capture stages of the sample preparations performed for single-molecule fluorescence and cryo-TEM microscopy. Rachid Sougrat of the Imaging and Characterization Core Lab helps to analyze the results (bottom right).
© 2019 KAUST
The histone decorations in the DNA origami platform therefore greatly simplify the design principles of the nanotechnology, says Maged Serag, a research scientist in Habuchi’s lab. What’s more, he adds, “The fact that we integrated a protein in the overall structure helps increase the applicability of our approach in different aspects of the biotechnology field.”
Habuchi, Serag and their coworkers have been working to advance the technology.
“We are trying to integrate histone proteins at specific locations within DNA-origami structures, a first step toward constructing complicated nanostructures,” Serag explains.
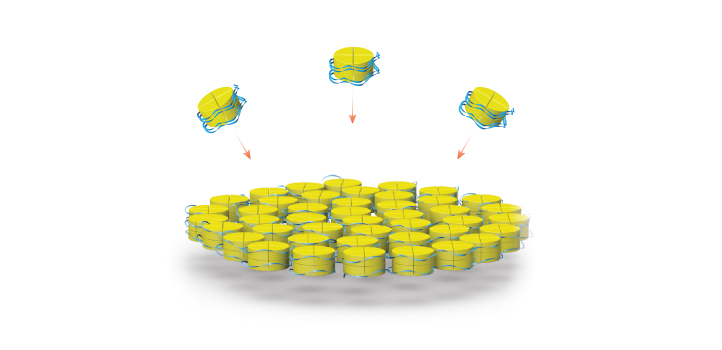
Histones and DNA self-assemble into nanoparticles and cross-linked complexes.
© 2019 KAUST; Heno Hwang
The hybrid assemblies could also help scientists better understand the basic role of histone proteins in regulating gene expression and DNA replication. The study, Habuchi says, could shed light on these fundamental biological functions.
References
-
Serag, M.F., Aikeremu, A., Tsukamoto, R., Piwoński, H., Abadi, M., Kaji, N., Dwyer, J.R., Baba, Y. & Habuchi, S. Geometry-based self-assembly of histone–DNA nanostructures at single-nucleotide resolution. ACS Nano 13, 8155–8168 (2019).| article
You might also like

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience
AI speeds up human embryo model research
Bioscience




