Computer Science
Database to support infectious disease research
Linking disease pathogens to clinical signs and symptoms through a database could support research into the molecular mechanisms of infectious diseases.

A new database PathoPhenoDB facilitates the search for associations between infectious diseases, the pathogens that cause them, the resulting clinical signs and symptoms, and the drugs that can treat them. It also contains information on the proteins and genetic changes that can make pathogens resistant to treatment with certain drugs.
Developed by researchers at KAUST in collaboration with the University of Cambridge, UK, PathoPhenoDB can help infectious disease diagnosis and treatment, in addition to the study of the molecular mechanisms behind pathogen-host interactions.
KAUST scientists are combining computations, clinical and biological expertise to link disease pathogens to clinical symptoms.
© 2019 KAUST
“Several databases already exist for infectious disease research,” says KAUST research scientist, Şenay Kafkas. “But these either cover part of the data available in our database, like including only disease–pathogen associations, or they focus on different aspects of the diseases, like host-pathogen interactions for understanding the disease mechanism.”
To put together a comprehensive database, the team collected data from existing databases and then used artificial intelligence to automatically identify more information from the literature, explains Kafkas. The team’s clinical geneticist, Marwa Abdelhakim, then manually combed through the data for an accuracy check, adding or removing information where needed.
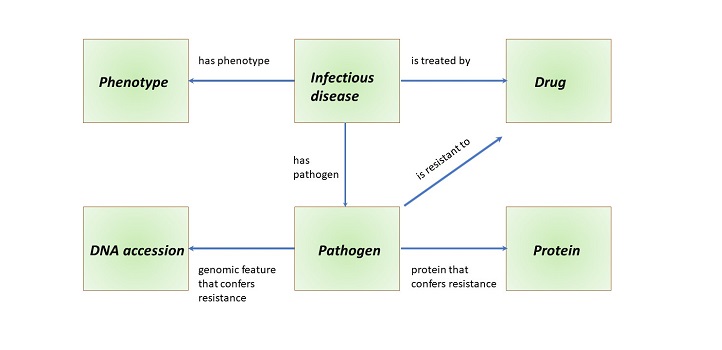
Schematic overview of the types of entities and their relationships to each other in the databased PathoPhenDB.
Reproduced from reference one under a creative commons license © JScientific Data 6, © 2019 / CC BY 4.0
“The group’s combination of computational, clinical and biological expertise put it in a strong position to generate PathoPhenoDB,” says Paul Schofield, a Reader in biomedical informatics at the University of Cambridge.
PathoPhenoDB is unique in that it links pathogens to disease phenotypes: the clinical signs and symptoms of disease. “Phenotypes encode for the molecular and physiological mechanisms underlying disease and can therefore be used to study these mechanisms,” says KAUST’s Robert Hoenhdorf, who led the initiative.
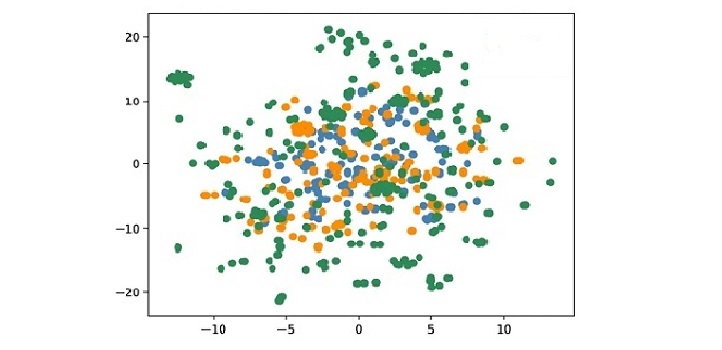
A plot from the database that shows the distribution of phenotypes elicited by pathogens. Viruses are colored in blue, bacteria in orange and all other pathogens in green.
Reproduced from reference one under a creative commons license © JScientific Data 6, © 2019 / CC BY 4.0
The database is publicly accessible and searchable from http://patho.phenomebrowser.net/. To use it, a term, which can be a pathogen, disease, or disease phenotype, is typed into the search box. The database brings up any information it holds that is associated with the search term.
PathoPhenoDB currently covers associations between 508 infectious diseases and 692 taxa of pathogens. It also includes information about drugs that can treat 130 infectious diseases and their associated pathogens. Finally, it includes information on known mechanisms of drug resistance for 30 pathogens.
“This will interest clinical infectious disease investigators and the bioinformatics community; particularly the latter as the database brings together data in a way that can be readily shared, integrated and computed upon,” says Schofield. “This mobilization of data, particularly from a manually validated and curated database, can provide inputs into many projects. We sow the seeds and wait for the flowers to grow!”
“PathoPhenoDB is a community resource and will evolve with the demands of the scientific community,” adds Kafkas.
The team plans to keep the database up to date through regular repeats of their automated workflows. It also aims to include next-generation sequencing data for pathogens, which could help improve infectious disease diagnosis and treatment.
References
- Kafkas, S., Abdelhakim, M., Hashish, Y. Kulmanov, M. Abdellatif, M. Schofield, P.N. & Hoehndorf, R. PathoPhenoDB, linking human pathogens to their phenotypes in support of infectious disease research. Scientific Data 6, 79 (2019).| article
You might also like

Computer Science
Green quantum computing takes to the skies

Computer Science
Probing the internet’s hidden middleboxes

Bioscience
AI speeds up human embryo model research
Computer Science
Improving chip design on every level

Computer Science
Sweat-sniffing sensor could make workouts smarter

Computer Science
A blindfold approach improves machine learning privacy
Computer Science
AI tool maps hidden links between diseases
Bioscience




