Bioscience
Global study links newly named syndrome with other disorders
Mutations in a repetitive protein motif are responsible for a rare cognitive disorder.

A new kind of congenital disorder caused by a group of mutations in a gene previously linked only to a rare progressive brain disease has been discovered by an international team co-led by Saudi investigators.
The disorder—congenital hypotonia, epilepsy, developmental delay, digit abnormalities, or CHEDDA—is characterized by intellectual disability, reduced muscle tone and facial abnormalities. It results from defects in ATN1, a gene also implicated in a neurodegenerative condition called dentatorubral-pallidoluysian atrophy (DRPLA). However, the disease-causing mutations responsible for each illness occur in different regions of the gene, and the clinical symptoms of DRPLA and CHEDDA are observably distinct.
The discovery stems from a collaboration between two clinical geneticists—Fowzan Alkuraya of King Faisal Specialist Hospital and Research Center in Saudi Arabia and Emma Palmer from Sydney Children’s Hospital in Australia—each of whom had a young patient with a mysterious condition suspected to arise from ATN1 mutations.
To understand the origin of the novel disorder, the clinicians contacted KAUST structural biologist, Stefan Arold and his group at the Computational Bioscience Research Center to analyze their patients’ mutations on a molecular level. Although the CHEDDA-affected children did not have the exact same genetic defect, Arold’s team found that all mutations disrupted the same stretch of the protein encoded by ATN1—one contained an unusual repetitive structure, which they named the HX repeat motif.
Follow-up studies conducted with KAUST teams led by cell biologist Jasmeen Merzaban and structural biologists Mariusz Jaremko and Łukasz Jaremko further uncovered the function of this enigmatic motif and how CHEDDA-related gene variants alter its molecular characteristics. The KAUST researchers also identified similar motifs encoded by a handful of other genes linked to cognitive disorders, suggesting that the team may have discovered a mutational mechanism of broad clinical significance.
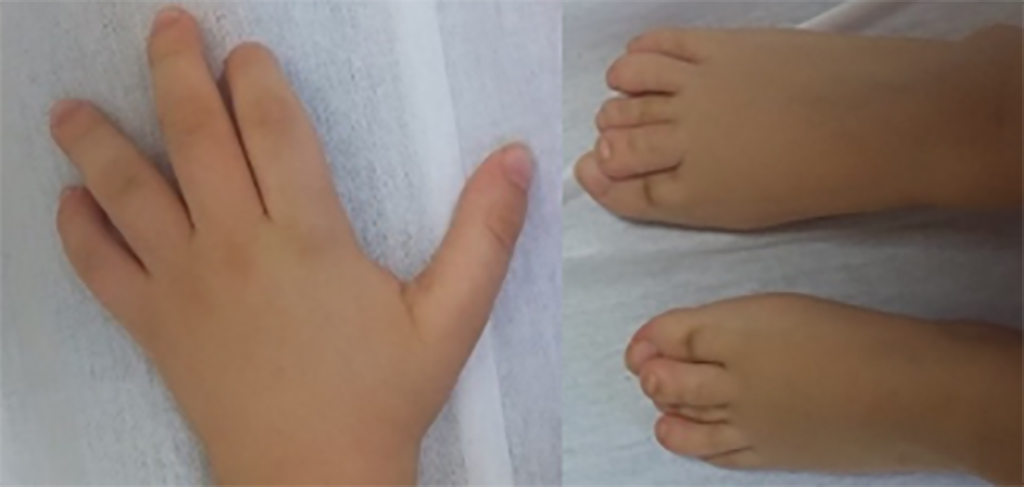
The hands and feet of patients with CHEDDA commonly have features that include abnormalities of the palmar creases, bulbous endings to the fingers and toes, and toes that overlap.
© Reproduced with permission from reference 1 © 2019 journal
While the laboratory experiments proceeded, doctors around the world began contacting Alkuraya and Palmer with other suspected cases of CHEDDA, and Arold confirmed that the ATN1 mutations of all patients disrupted the same HX repeat motif. Together, they ultimately assembled a cohort of eight children with the disorder between the ages of two months and nine years, the oldest being a Saudi child treated by Alkuraya.
By putting a name to a mysterious illness and revealing its genetic underpinnings, fewer families should now have to endure harrowing diagnostic odysseys, Palmer says. The finding also helps bring access to much-needed medical care, genetic counseling and specialist support. And importantly, parents of affected children can now communicate with each other, sharing their experiences and forming support groups. “That is very rewarding for me,” she says.
For researchers like Arold, the description of CHEDDA is also the first step in the long process of developing new treatments for affected children. “As we continue unraveling the cellular and molecular basis of this condition,” Arold says, “we will be in a better position to direct treatment and therapy.”
References
- Palmer, E.E., Hong, S., Al Zahrani, F., Hashem, M.O., Aleisa, F.A…. Arold, S. De novo variants disrupting the HX repeat motif of ATN1 cause a recognizable non-progressive neurocognitive syndrome. American Journal of Human Genetics 104, 542–552 (2019).| article
You might also like

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience
AI speeds up human embryo model research
Bioscience




