Bioscience
New moves for polymer chain dynamics
A Nobel-winning method for observing single molecules in action provides the basis for unprecedented insights into polymer chain dynamics.
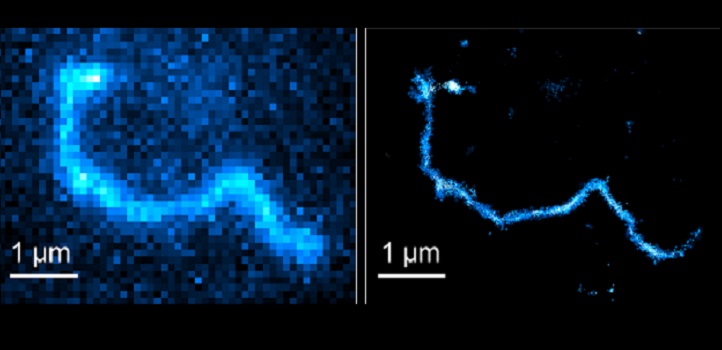
A technique that enables researchers to watch the motion of individual molecules within a polymer has been developed by KAUST: it challenges current thinking about polymer physics and could lead to new materials that can be tailored for specific tasks.
Polymers are a large and diverse family of materials ranging from hard, rigid plastics to flexible, stretchy gels. At the microscopic level, polymers consist of long-chain molecules that are tangled together like a nanoscale plate of spaghetti. The properties of a polymer material arise from the way its component polymer chains move and interact with each other. Until now, researchers’ ability to fully understand polymer properties was hampered because it was impossible to observe individual polymer chain motion.
Satoshi Habuchi and his team has overcome this limitation using super-resolution fluorescence microscopy. “Fluorescence imaging is an excellent technique to capture real-time behavior of dynamic systems,” says Maram Abadi, a member of Habuchi’s team.

A new technique developed by Maram Abadi (left), Satoshi Habuchi and colleagues challenges current thinking about polymer physics.
© 2018 KAUST
For the polymer study, Habuchi and his team created a polymer with fluorescent tags attached at several points along the chain. Although the spatial resolution of conventional fluorescence imaging is limited to 200–300 nanometers—insufficient for tracking polymer chain dynamics —super-resolution fluorescence imaging offers considerably sharper 10–20 nanometer resolution. Super-resolution is achieved by capturing 10,000 separate fluorescence microscopy images within a few seconds, and then using a computer to combine them to generate a single super-resolution image. The technique earned its original discoverers the Nobel Prize in Chemistry 2014.
Habuchi and his team combined this technique with a single-molecule tracking algorithm they recently developed. “It provided a powerful tool for investigating entangled polymer dynamics at the single-molecule level, Abadi says.
The tool showed polymer dynamics are more complex than previously thought. Polymer dynamics has been modeled using reptation theory in which the entire polymer chain is considered to move as a single unit, similar to a snake, which explains the term’s derivation from the word reptile. Super-resolution fluorescent microscopy reveals that the polymer actually undergoes chain-position-dependent motion, with most motion occurring at the chain ends and the least motion occurring in the middle.
This discovery shows that polymer physics theory will have to be revised, Abadi says. “Since rheological properties of materials arise microscopically from entangled polymer dynamics, a revision of the reptation theory would have a broad impact not only on fundamental polymer physics but also on the development of a wide range of polymer nanomaterials,” she says.
The team now plans to apply its technique to more complex systems, including polymer gels and networks of biomolecules within cells.
References
- Abadi, M., Serag, M.F. & Habuchi, S. Entangled polymer dynamics beyond reptation. Nature Communications 9, 5098 (2018).| article
You might also like

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience
AI speeds up human embryo model research
Bioscience




