Bioscience
Fastening enzyme seals the deal in genome repair
Structural biology study reveals dynamics of DNA ligation during genome replication.
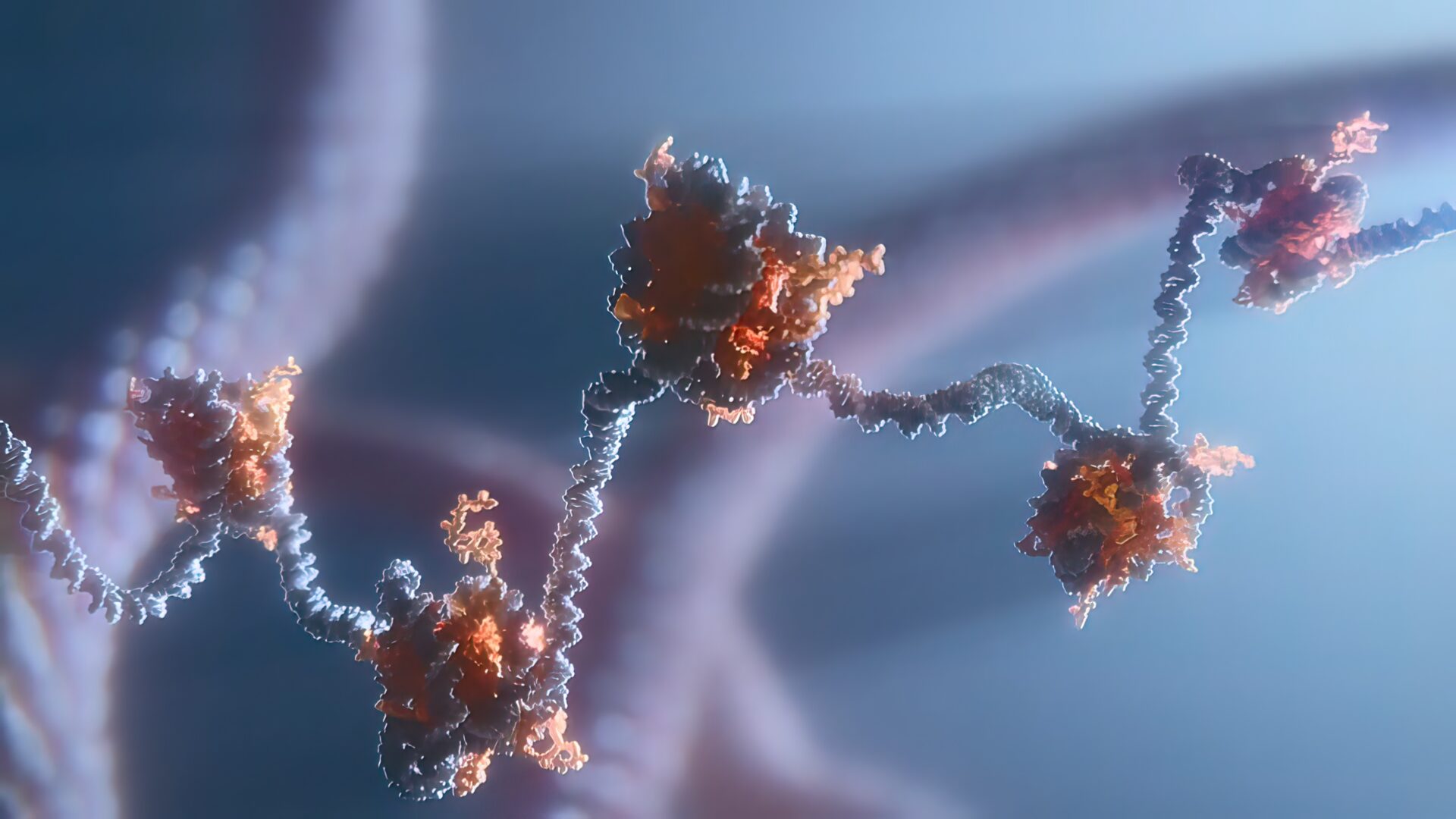
Unconnected strands of DNA must be sealed together during genome replication and repair, and a new KAUST-led investigation shows how the fastening enzyme involved gets the job done.
By combining cryogenic electron microscopy (cryo-EM) imaging, structural modeling and biochemical assays, the researchers have shown how this enzyme, known as human ligase 1 (Lig1), quickly scans the genome looking for nicks in DNA. To guide the search, Lig1 hitches itself to a ring-shaped protein called PCNA that encircles strands of genetic material and travels along the DNA like a sliding clamp. When a nick is found, Lig1 changes its conformation and wraps around the DNA to seal the nick.
The interaction between Lig1 and PCNA in turn dislodges another enzyme, a protein called FEN1. As the researchers demonstrate, this ouster of FEN1 is necessary to prepare the nicked DNA before Lig1 can clinch it. “This is an essential process that occurs every time one of two strands of the DNA double helix experiences a break,” says structural biologist Alfredo De Biasio, who co-led the study with his KAUST colleague Samir Hamdan. “By using cryo-EM,” De Biasio continues, “we showed that PCNA functions as a kind of toolbelt to which both enzymes can attach independently and simultaneously, enabling the efficient handoff of the nicked DNA from FEN1 to Lig1 and the sealing of the nick.” This molecular handoff between enzymatic tools on the PCNA toolbelt underpins various cellular processes, including DNA repair and recombination.
To dissect the toolbelt’s structure, however, the researchers focused on just one process: DNA copying — specifically the synthesis and conjoining of short DNA fragments created during part of genome replication. PCNA’s recruitment of the nick-sealing enzyme relies on a delicate dance between different parts of Lig1. First, Lig1 tethers its front end to the PCNA ring near sites of discontinuities in the genome. Lig1 then swivels around, encircling the DNA and contacting PCNA through the so-called DNA binding domain, ultimately forming a two-ringed stack structure around the nicked DNA. This conformational change leads to the displacement of FEN1, an enzyme that cuts away extraneous flaps of DNA generated at certain stages of genomic replication and repair. With FEN1 out of the picture, Lig1 is free to circle around the DNA at the nick, an essential step for Lig1 to fulfill its DNA-fastening function, as experiments with a mutated form of the enzyme showed. The findings are the latest in a series of papers from De Biasio and Hamdan — a fruitful collaboration between the two scientists, and their lab groups have helped reveal the structural mechanics of human DNA repair and replication.
References
- Blair, K., Tehseen, M., Raducanu, V.-S., Shahid, T., Lancey, C., Rashid, F., Creuhet, R., Hamdan, S.M. & De Biasio, A. Mechanism of human Lig1 regulation by PCNA in Okazaki fragment sealing. Nature Communications 13, 7833 (2022).| article
You might also like

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience
AI speeds up human embryo model research
Bioscience




