Bioscience
Old methods prove true for studying proteins
A decades-old technique for probing protein motions proves more accurate than current practices.
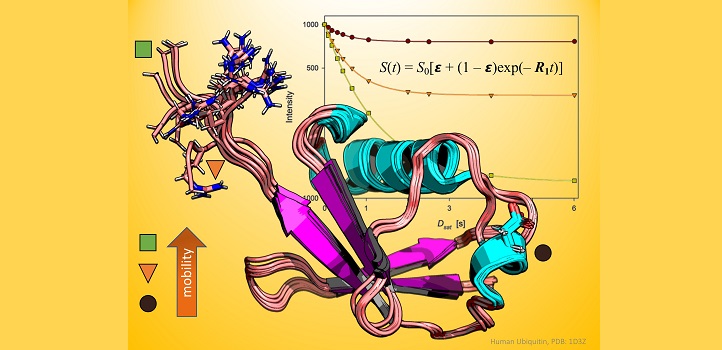
A fresh new look at an old technique in protein biochemistry has shown that it should be reintroduced to the spectroscopy toolkit.
For decades, scientists have used nuclear magnetic resonance (NMR) spectroscopy to probe the molecular motions of proteins on various timescales. This technique has revealed aspects of enzyme reactions, protein folding and other biological processes, all on an atomic scale.
Typically, spectroscopists will gauge the rotation of NMR-active atoms in the protein backbone with and without proton irradiation to calculate a ratio known as a steady-state nuclear Overhauser effect (NOE); however, it was not always done this way.
Before steady-state NOE experiments became the norm in biological investigations, scientists would often take a greater number of measurements over the course of an irradiation experiment. This method, termed “dynamic” NOE, might seem more complicated, but according to Ph.D. student Vladlena Kharchenko, it is no more time consuming than steady-state NOE, while dynamic NOE provides additional information about protein flexibility and is far more accurate to minute biological motions in proteins.
“It works for proteins and makes studying their dynamics even more accurate,” says Kharchenko, a member of Łukasz Jaremko’s lab at KAUST. “Our message to biological NMR spectroscopists is simple: ‘Don’t be afraid of dynamic NOE.’”
To prove the technique’s worth, Kharchenko, Jaremko and their team performed a series of NMR experiments on ubiquitin, a globular protein that regulates a range of processes inside the cell. Working with Mariusz Jaremko, also from KAUST, and collaborators in Poland, the researchers collected both steady-state and dynamic NOE measurements and demonstrated that the dynamic approach is always preferable—except under a few specific conditions, such as when instrument access is limited or when proteins degrade very rapidly.
Notably, the steady-state approach proved especially prone to errors in regions of the ubiquitin protein that were flexible and disposed to moving around. The dynamic technique, in comparison, offered no such misleading results.
In light of their findings, the KAUST team hopes that other scientists with an interest in atomic-level protein mechanics will now begin to adopt, or at least reconsider, dynamic NMR methods. Kharchenko says that sometimes, “it’s worth dusting off forgotten methods and checking if they fit to new emerging questions and systems of research interest.”
References
-
Kharchenko, V., Nowakowski, M., Jaremko, M., Ejchart, A. & Jaremko, L. Dynamic 15N{1H} NOE measurements: a tool for studying protein dynamics. Journal of Biomolecular NMR 74, 707-716 (2020).| article
You might also like

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience
AI speeds up human embryo model research
Bioscience




