Statistics
Clustering helps unlock secrets of the human brain
The latest statistical methods from research on complex high-dimensional environmental data also yield powerful tools for interpreting brain activity.
Environmental science and neuroscience may seem poles apart as research endeavors, but both are underpinned by the need to analyze and interpret enormous datasets capturing complex spatio-temporal processes. Statistically, looking for patterns and relationships in such datasets is very similar, whether it’s measurements of temperature across the globe or electrical activity throughout the brain. This common purpose has brought together Ying Sun and Hernando Ombao—two of KAUST’s leading researchers in big data statistics.
It all starts with the weather
In environmental monitoring data, each meteorological parameter—temperature, wind speed or precipitation—and each measurement station represents a dimension of the consolidated dataset. The result is a very large dataset with a complexity that defies conventional analytical approaches.
“We focus on developing new statistical methods for analyzing the complex high-dimensional data typically encountered in environmental science,” says Sun.
Sun and her team have proposed a suite of new statistical approaches for dealing with this data, including highly flexible and computationally efficient methods for dealing with very large datasets.
Into the brain
Inspired by problems in neuroimaging, Ombao’s group have been developing similar statistical tools to better understand the relationships and dependences among spatio-temporal signals. Techniques, such as functional magnetic resonance imaging (fMRI) and electroencephalography (EEG), that capture different aspects of brain activity in time and space with high dimensionality are similar in many ways to the type of environmental data being worked on by Sun’s team; as such, many of the same statistical approaches apply.
“Our major focus is on understanding the role of brain connectivity and its associations with mental and neurological diseases,” says Ombao. “When looking at brain activity, different regions are activated as a person processes information, and some regions respond in an organized or synchronized manner. The goal of our recent work has been to develop a new statistical clustering method that identifies brain regions with synchronous behavior and discover common features and group patterns among brain signals that could help us understand brain functional connectivity.”
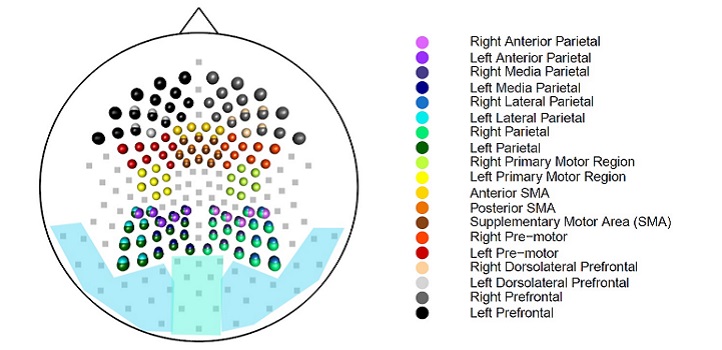
This visualization shows the evolving connectivity of the left temporal brain (T3) region. The white node shows the location of the T3 channel. Blue nodes indicate no connectivity and red nodes indicate connectivity in the region.
© 2019 Marco Pinto
Bringing it together
According to Ombao, the biggest challenge when applying clustering methods to brain signal data is how to define the features of the time series, and then how to quantify their similarity. The team’s research considers two different measures of similarity to identify clusters—spectral synchronicity and cluster coherence. These led to the development of hierarchical clustering algorithms for EEG data.
“One way to study functional connectivity in the brain is to look for similar patterns of activation in different regions,” says Ombao. “Modern EEG technologies allow us to record data every millisecond across hundreds of channels, meaning that a recording of even a few minutes can result in a very large dataset. To analyze such datasets more effectively, we have developed two clustering algorithms that are computationally fast and provide an accurate and interpretable summary of brain-region connectivity.”
EEG signals are commonly studied by analyzing their frequency composition, akin to picking out the harmonics that give different musical instruments their distinct sound. A high degree of similarity in frequency composition could mean that two signals are functionally connected. Postdoctoral fellow, Carolina Euan, in collaboration with Ombao and Joaquín Ortega, from the Center for Mathematical Investigation in Mexico, developed the hierarchical spectral merger clustering method to quickly identify groups of similar signals with discrete frequency bands1. However, these clusters are not necessarily dependent in a functional connectivity sense. To refine the analysis, Euan, Ombao and Sun worked together on a hierarchical cluster coherence method to identify those clusters that are highly dependent within specific frequency bands2.
“By applying our method to EEG data, we can pick out brain regions that are interdependent and identify the underlying frequency band in which they are functionally synchronized,” says Euan.
What’s next?
“So far, we have only considered one subject at a time,” says Ombao. “Next we plan to model clustering variability between different test subjects, as well as understand the evolution of clustering across a range of stimuli. This approach could also be useful for comparing brain-network clustering in healthy subjects and patients with a brain disease.”
References
You might also like

Statistics
Checking your assumptions
Statistics
Internet searches offer early warnings of disease outbreaks
Statistics
Joining the dots for better health surveillance

Statistics
Easing the generation and storage of climate data
Statistics
A high-resolution boost for global climate modeling

Applied Mathematics and Computational Sciences
Finer forecasting to improve public health planning

Bioengineering
Shuffling the deck for privacy
Bioengineering




