Material Science and Engineering
DNA imaging made easy
New method offers a means of efficient and high-throughput technique to study the structure of DNA.
It may be the most famous structure in biology, but it wasn’t until a few years ago that biophysicist Enzo Di Fabrizio and his colleagues took the first direct images of the DNA double helix with an electron microscope.
Now, Di Fabrizio and his lab group at KAUST have improved upon their groundbreaking technique, tweaking the protocol to make it simpler and faster.
“It’s an efficient and high-throughput alternative to more conventional techniques,” says Monica Marini, postdoctoral fellow in Di Fabrizio’s lab and first author of the new study.
The image that helped Watson and Crick decipher the corkscrew-like structure of DNA 65 years ago was taken using a technique called X-ray crystallography, which involves scattering electromagnetic radiation off atoms in a crystalized form of DNA. For decades, this was the only way to get 3D renderings of the building blocks of life.
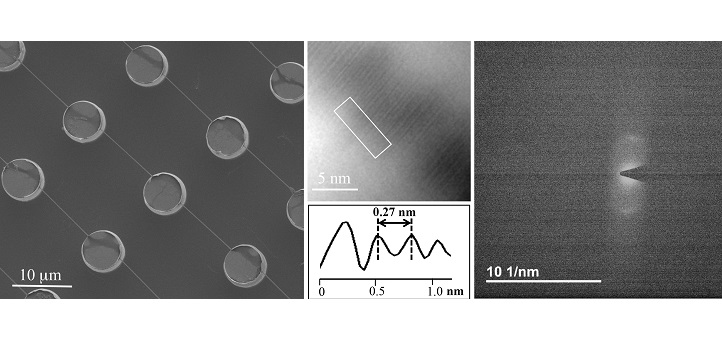
But those pictures were only abstractions, based on interpretations of diffracted X-rays. They weren’t true photographs. And it wasn’t until 2012—when Di Fabrizio, back in his native Italy, produced the first direct image of DNA—that researchers got a faithful picture of the double helix.
Di Fabrizio moved to KAUST in 2013, and over the past five years his group has been steadily improving and building upon the original imaging protocol, which involved transmission electron microscopy (TEM), a technique in which electrons are beamed onto photographic film.
Their method involves spreading minute droplets of fluid containing DNA onto silicon wafers etched with tiny cylindrical pillars and holes. As the droplets dry out, the DNA stretches itself out across the microscopic bed of pillars, creating spools of interconnected thread.
Previously, Di Fabrizio’s team has applied TEM to capture images of the DNA strands directly. But now, they’ve also performed a more straightforward diffraction analysis of the TEM beams—thus “creating an experiment that’s quite similar, in terms of physical principles, to that done by the scientists who first discovered the DNA structure,” says Andrea Falqui, another KAUST faculty member who has collaborated with Di Fabrizio in previous works.
As Di Fabrizio and Marini show, this diffraction-based approach produced pictures that measured the distance between rungs of the DNA ladder just as accurately as direct TEM imaging. Now, the researchers plan to use this technique to image more complex arrangements of DNA. For example, they want to look at DNA when it’s interacting with proteins, drugs or heavy metals.
All these interactions “can cause variations to the pristine conformation of the double helix,” Marini says, and soon they should have the photographic evidence to see those changes in detail.
References
-
Marini, M., Allione, M., Lopatin, S., Moretti, M., Giugni, A., Torre, B. & di Fabrizio, E. Suspended DNA structural characterization by TEM diffraction. Microelectronic Engineering 187–188, 39–42 (2018).| article
You might also like

Chemistry
Turning infrared solar photons into hydrogen fuel

Applied Physics
A single additive enables long-life, high-voltage sodium batteries

Bioengineering
Smart patch detects allergies before symptoms strike

Applied Physics
Two-dimensional altermagnets could power waste heat recovery

Applied Physics
Interface engineering unlocks efficient, stable solar cells

Applied Physics
The right salt supercharges battery lifespan

Applied Physics
Light-powered ‘smart vision’ memories take a leap forward

Applied Physics




