Bioscience
Enzyme from briny deep resurrected in the lab
Genomes of single microbial cells isolated from the Red Sea could yield a goldmine for biotechnology.
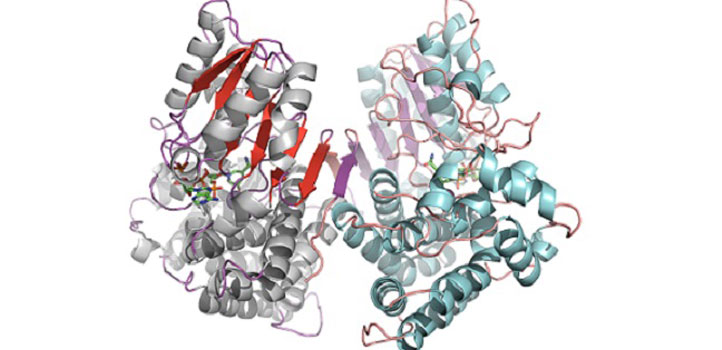
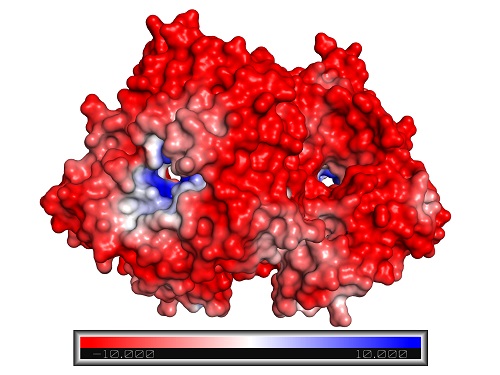
Closed configuration of the ADH/D1 dimer (same orientation as the figure above).
Reproduced with permission from reference 1© 2017 American Chemical Society
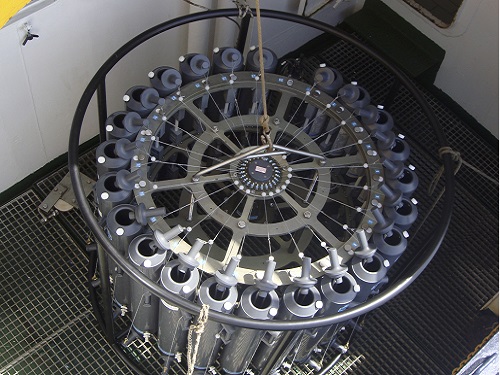
The rosette sampler is used to collect the bacterial samples from the briny pool in the Red Sea
© 2017 André Antunes, Edge Hill University, UK.
Mysterious microbes that thrive in hot and super-salty brine lakes at the bottom of the Red Sea could yield a treasure trove of new enzymes for industrial applications—if only scientists had access to their biological bounty.
A new study led by KAUST scientists now spells out a way to exploit this vast untapped resource.
Current methods rely on growing microbes in the lab to study their characteristics. However, most microbes cannot be cultured and thus evade scientific scrutiny. Instead, researchers from KAUST and the Technical University of Munich (TUM) in Germany resurrected and tested specific proteins from so-called single-amplified genomes (SAGs)—whole genomes extracted from a single captured microbial cell.
“This is the first time that SAGs have been used to produce proteins,” says first author, Stefan Grötzinger, a doctoral student working at both KAUST and TUM. “The proof that proteins of desired functions can be obtained from SAGs could change the way we search for new enzymes.”
Grötzinger and his colleagues—led by KAUST structural biologist Stefan Arold with Jörg Eppinger, a chemist formerly at KAUST, and TUM scientists Dirk Weuster-Botz and Michael Groll—started with a microbial cell plumbed from a brine pool located 80km off the coast of Jeddah and 2,000m below the Red Sea surface. From its SAG, they computationally identified a gene encoding one of the microbe’s alcohol dehydrogenases (ADHs), an enzyme commonly used in food, pharmaceutical and chemical industries.
The researchers first tried to express this enzyme in “Escherichia coli”, a common bacterial platform for protein production, but this approach didn’t produce useful proteins. They then turned to a different microbe that lives in a highly saline environment and that can also be cultured in the lab. In this microbe, they managed to obtain enough of the ADH protein that they could infer its three-dimensional structure through X-ray crystallography and perform a full biochemical characterization, including its enzymatic capacities.
Their analyses revealed characteristics that presumably arose as adaptations to life in the hot and salty sea. For example, the protein works under extremely high concentrations of organic solvent, tolerates high temperatures and can be freeze-dried—all features that make the enzyme attractive for commercial industrial applications, Grötzinger says.
But more generally, he adds, the study provides a roadmap for how to mine the molecular riches of organisms found in extreme environments. Plus, it provides an exemplar of international and local collaboration, uniting scientists in Germany and Saudi Arabia, with cooperation from three units of KAUST: the Biological and Environmental Science and Engineering Division, the Computational Bioscience Research Center and the Catalysis Center.
References
- Groetsinger, S., Karan, R., Strillinger, E., Bader, S., Frank, A., Al Rowaihi, I., Wackerow, W., Akal, A., Archer, J., Rueping, M., Weuster-Botz, D., Groll, M., Eppinger, J., & Arold, S. Identification and experimental characterization of an extremophilic brine pool alcohol dehydrogenase from single amplified genomes. ACS Chemical Biology 13, 161-170 (2018).| article
You might also like

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience
AI speeds up human embryo model research
Bioscience




