Material Science and Engineering
Silver nanospheres expose cancer’s single flaw
Arrays of tiny silver nanolenses can detect a unique mutation with the potential to cause cancer among a mix of human peptides.
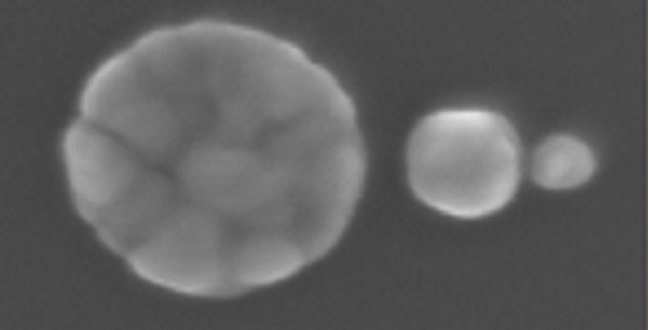
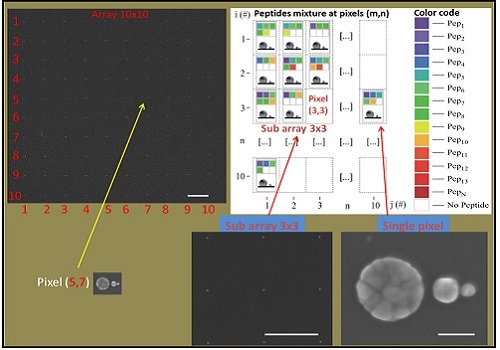
Top left: the SEM picture of 10×10 array of SSC; each SSC represents a pixel element of the matrix (the bar is 2 mm). In each pixel, a color code (see right) is associated to specific peptide. The bottom shows a sub matrix 3×3 pixels (the bar is 2 mm). Bottom right: a detailed SEM picture of the SSC representing the pixel (the bar is 50 nm). The empty pixels represent specific positions in the array where no peptides are detected.
© 2015 KAUST
Identifying molecular markers that predict potential tumor growth is a promising way to screen for diseases such as breast cancer. A KAUST-led international research team has developed a nanostructured sensor that analyses breast cancer samples and spots single point mutations in tumor suppressor genes with part-per-trillion sensitivity1.
The new device is based on ordered arrangements of silver nanospheres known as self-similar chains (SSCs). By tweaking the size and spacing of SSCs deposited onto surfaces, researchers can create “hotspots” where plasmons — quantized oscillations of electron density — are strongly brought into focus through geometric effects. The confined plasmons enhance the capabilities of techniques such as vibrational Raman spectroscopy so that few- or single-molecule detection becomes possible.
However, identifying a biomolecular cancer signature in a system as complex as a protein is challenging. For example, in the tumor-fighting BRCA1 protein, a single mutated amino acid on a peptide can induce hereditary predispositions to breast and ovarian cancers. Any Raman spectra associated with the mutated peptide is washed out by other molecular groups in the protein.
Enzo Di Fabrizio from KAUST and colleagues aimed to improve Raman-based biosensing by arranging a matrix of SSCs into a 10×10 grid pattern. Under this method, each SSC acts as a nanoscale “pixel”that captures Raman spectra of molecules sitting in the hotspots. By calibrating the hotspot signals using serial dilutions, individual peptides residing inside complex mixtures can be identified.
Putting this concept into practice called for the manufacture of silver SSCs with extreme precision: optimal hotspots appeared between spheres separated by less than 5 nanometers.
“We used a combination of top-down and bottom-up approaches in this fabrication,” explained Di Fabrizio.”High resolution electron beam lithography allowed the best control of structure positioning, while selective reduction of silver ions created self-assembled 3D nanostructures of appropriate size and shape.”
The researchers used enzyme digestion technology to reduce BRCA1 into a mix of 25 different peptides for testing. After depositing sample droplets on the SSC matrix and performing Raman spectroscopy, they compared the pixel signals to a database of peptide Raman spectra with a fitting algorithm. This procedure yielded pixel-by-pixel information of the mixture’s makeup, including the distinct signature of the mutated peptide.
Di Fabrizio noted that this strategy, which can also be applied to chemotherapy and drug response measurements, owes its success to stronger-than-expected Raman signals arising from surprising surface roughness contributions. “The roughness enhances the electric field and improves sensitivity by an order of magnitude,” he said.
References
-
Coluccio, M.,L., Gentile, F., Das, G., Nicastri, A., Perri, A.M. et al. Detection of single amino acid mutation in human breast cancer by disordered plasmonic self-similar chain. Science Advances 1:e1500487 (2015). | article
You might also like

Chemistry
Turning infrared solar photons into hydrogen fuel

Applied Physics
A single additive enables long-life, high-voltage sodium batteries

Bioengineering
Smart patch detects allergies before symptoms strike

Applied Physics
Two-dimensional altermagnets could power waste heat recovery

Applied Physics
Interface engineering unlocks efficient, stable solar cells

Applied Physics
The right salt supercharges battery lifespan

Applied Physics
Light-powered ‘smart vision’ memories take a leap forward

Applied Physics



