Bioscience
Molecular movie reveals enzyme’s mechanism of action
An enzyme involved in maintaining genome fidelity goes through seven steps to recognize its DNA substrates.
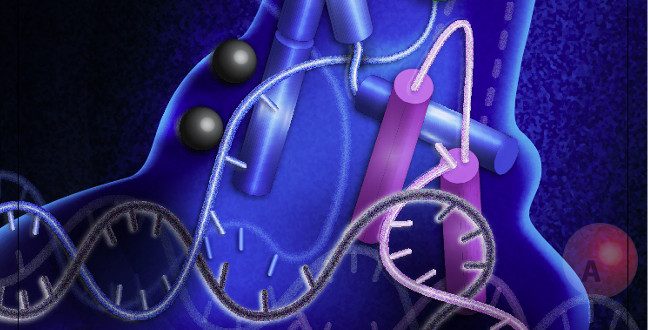
DNA replication and repair are essential processes for all living cells, but key details about the 5′ nucleases, a family of enzymes central to these processes, remain a fundamental mystery. Using a sophisticated imaging method, researchers from KAUST captured molecular movies of one such enzyme — a central component of cellular DNA metabolism called human flap endonuclease (hFEN1) — and showed the steps it takes to recognize and process its substrate1.
The findings add to a basic understanding of genome fidelity as well as offer a possible new strategy for combating cancer. “hFEN1 is highly expressed in proliferating cells in a manner that correlates with tumor aggressiveness,” says Samir Hamdan, an associate professor in KAUST’s Biological and Environmental Science and Engineering Division, who led the research. Hamdam explained that it was important to understand the mechanism by which 5’ nucleases recognize and cleave their substrates, as this could pave the way for a broad-spectrum anticancer drug that targets members of 5’ nucleases such as hFEN1.
Scientists already knew that hFEN1 removes overhanging segments of single-stranded DNA that form when the cell is repairing genetic damage or copying the genome. It does this partly by grasping and severely bending DNA before trimming the ‘flap’, but the mechanism of its action was poorly understood. To remedy this, Hamdan and his colleagues characterized the dynamics of hFEN1 down to the single molecule level.
With the help of a fluorescence imaging technique known as Förster resonance energy transfer, Hamdan’s team at KAUST, led by postdoctoral fellow Mohamed Sobhy, found that this bending occurs only after hFEN1 has completed a series of seven intermediary steps to recognize the DNA strands it will eventually slice and dice.
“We found that DNA bending is a highly specific and complex intermediary step that is induced only after hFEN1 thoroughly verifies all the substrate features,” explains Hamdan.
One problem though, according to Hamdan, is that the close structural similarity among 5’ nucleases makes it difficult to design a drug that would be specific for hFEN1 or other enzymes in the same family without potentially dangerous unintended side effects.
But Hamdan already has a proposal for how his group can puzzle their way to overcome this hurdle. “My research group is using powerful single-molecule imaging approaches to characterize different members of the 5′ nuclease superfamily.” says Hamdan. “This would provide a mechanism to identify common elements as well as individual characteristics that allow specific substrate recognition and so help to address this problem.”
References
- Sobhy, M.A., Joudeh, L.I., Huang, X., Takahashi, M. & Hamdan, S.M. Sequential and multistep substrate interrogation provides the scaffold for specificity in human flap endonuclease 1. Cell Reports 3, 1785–1794 (2013).| article
You might also like

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience
AI speeds up human embryo model research
Bioscience




