Bioscience
The long and short of DNA replication
An unexpected two-step mechanism occurs when cells copy DNA.

The process of copying DNA is complex choreography that requires rapid speeds and pin-point precision. Discovering the intricate details of this process could identify new ways to target diseases, such as cancer, and KAUST scientists have just uncovered a crucial step.
The DNA in each human cell is around 3 billion digits long and has to be copied every time a cell divides—which occurs nearly 2 trillion times each day.
If errors occur in DNA replication, cells can become abnormal and give rise to disease. DNA code that has mutated to allow the cell to divide uncontrollably, for instance, can lead to cancer. This makes it crucial to understand how enzymes replicate and repair DNA, says Manal Zaher, Ph.D. student in Samir Hamdan’s group at KAUST and first author of the study.
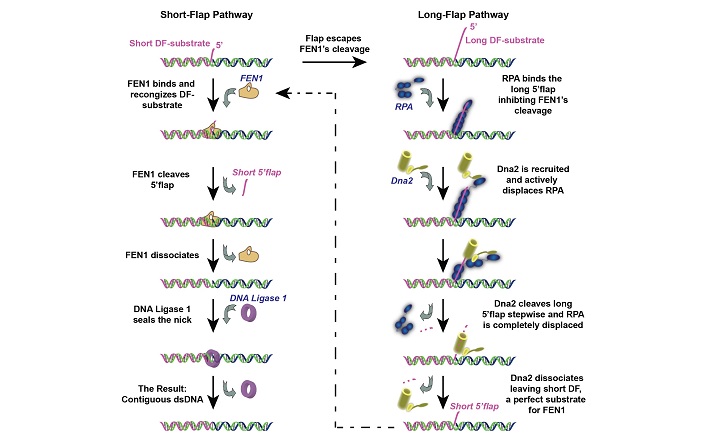
Zaher investigated the enzymes that allow short DNA strands formed during DNA replication, called Okazaki fragments, to link up into a continuous string of DNA.
By a process known as Okazaki fragment maturation, an enzyme called FEN1 cuts off “flaps” in the Okazaki fragments that need to be removed before the fragments can join up. “Sequence-based specificity partially explains the secret of replication fidelity; however, we lack key information about the basis for the structure-based excision by FEN1 required at ~50 million Okazaki fragment sites during human DNA replication,” Hamdan explains.
FEN1 is overproduced or altered in many forms of cancer, and Zaher wanted to investigate the exact involvement of this enzyme.
The researchers used an imaging technique called single-molecule FRET to discover how FEN1 docks and is released from other enzymes and DNA fragments.
KAUST scientists discovered that the enzyme FEN1 acts as a switch during DNA replication.
Reproduced from reference 1 under a Creative Commons Attribution license.© 2018
Zaher was surprised to discover that FEN1 acted as a switch between two different enzyme pathways. If the flap was short, FEN1 docked to the DNA to let the Okazaki fragments join. However, if the flap was long, FEN1 bounced off, leaving two other enzymes called Dna2 and RPA to dock on and cut the bulk of the flap before FEN1 in turn came in to finish the job.
“This two-step mechanism could significantly help our understanding of the regulation of the subsequent Okazaki fragment steps,” explains Zaher, “and it is important particularly because mutations in both Dna2 and RPA have also been linked to cancer.”
Hamdan’s group has set its sights on investigating the next interaction that FEN1 undergoes, continuing to unravel the mysteries of DNA replication one step at a time.
References
- Zaher, M.S., Rashid, F., Song, B., Joudeh, L.I., Sobhy, M.A., Tehseen, M., Hingorani, M.M. & Hamdan, S.M. Missed cleavage opportunities by FEN1 lead to Okazaki fragment maturation via the long-flap pathway. Nucleic Acids Research 6, 2956-2974 (2018).| article
You might also like

Bioengineering
Bio-inspired network structures for next-generation AI

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience



