Bioscience
A terminator for truncated transcripts
A gene linked to plant stress response helps to prevent the production of defective proteins.
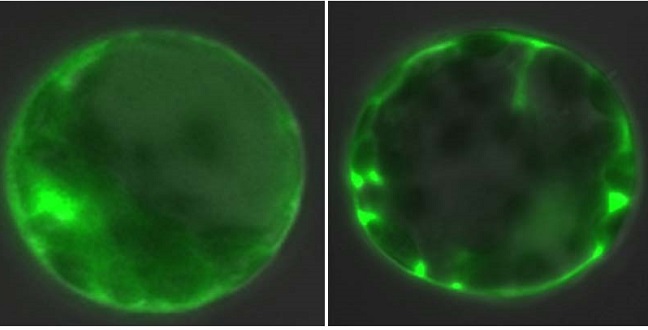
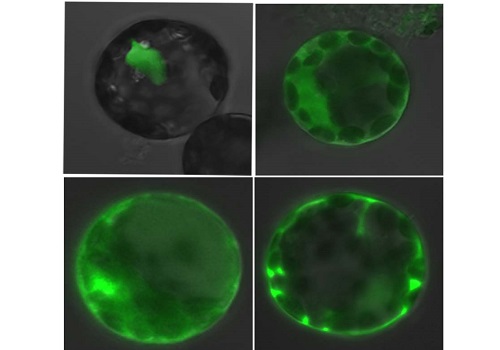
The top row shows that fluorescently labeled eIF4AIII localizes in the nucleus of normal Arabidopsis cells (left), but shifts to the cytoplasm in mutants lacking FRY2 (right). The bottom row shows that treatment with a chemical that inhibits FRY2 has a similar effect on eIF4AIII localization in both normal and mutant cells.
Reproduced with permission from reference 1 © 2016 Copyright American Society of Plant Biologists
Cells must be able to recognize and eliminate defective RNA transcripts before they can be translated into abnormal and potentially damaging proteins. Researchers led by Liming Xiong from KAUST have now identified an important aspect of this “quality control” process1.
Xiong and colleagues set out to learn more about how plants recognize and adapt to stressful changes in the environment. Their experiments with mutant strains of the thale cress plant Arabidopsis thaliana led them to the gene FIERY2 (FRY2).
“These mutants had increased expression of certain stress-responsive genes,” said Xiong. “They showed increased tolerance to salt stress and the stress hormone abscisic acid during seed germination, but were also more sensitive to freezing damage at the seedling stage.”
However, the researchers also noticed other intriguing futures of their FRY2 mutant, fry2-1. This mutant of the FRY2 gene gives rise to messenger RNAs (mRNAs) that contain a “premature termination codon”—a signal that interrupts protein translation early, resulting in a truncated protein.
These defective mRNAs are usually eliminated by a process called nonsense-mediated decay (NMD), but the mutant plants were instead accumulating high levels of the mutated FRY2 mRNA.
“We thus hypothesized that the FRY2 gene might be involved in NMD,” Xiong said.
The researchers gained further support for this model with experiments showing that the FRY2 protein interacts with eIF4AIII, a protein that helps anchor the NMD machinery to mRNAs slated for destruction. Over the course of this interaction, FRY2 chemically modifies eIF4AIII in a manner that ensures its correct localization within the cell. Absence of functional FRY2 causes eIF4AIII to accumulate in the cytoplasm rather than the nucleus, where it is usually found.
These results suggest guilt by association; however, other data offered more support for a direct link between FRY2 and NMD. Many genes routinely give rise to irregular “alternative” mRNA transcripts that are normally degraded via NMD, but the fry2-1 mutant plant cells instead accumulated these transcripts.
Intriguingly, chemical treatments that interfered with FRY2’s ability to chemically modify eIF4AIII restored the NMD-mediated elimination of these transcripts in fry2-1 mutants, highlighting the importance of the interaction between these two proteins in regulating NMD.
In future studies, Xiong’s team will delve deeper into the functional importance of the FRY2-eIF4AIII partnership in regulating NMD.
“Since FRY2 is involved in stress response,” he added, “we will also investigate the relationship between NMD and plant stress tolerance.”
References
- Cui, P., Chen, T., Qin, T., Ding, F., Wang, Z. et al. The RNA polymerase II C-terminal domain phosphatase-like protein FIERY2/CPL1 interacts with eIF4AIII and is essential for nonsense-mediated mRNA decay in Arabidopsis. Plant Cell advance online publication, 17 February 2016 (doi: 10.1105/tpc.15.00771). | article
You might also like

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience
AI speeds up human embryo model research
Bioscience



