Computer Science
Unlocking complex interactions
Functions of interacting molecular complexes are uncovered through a novel method of aligning the interaction interface of each complex.
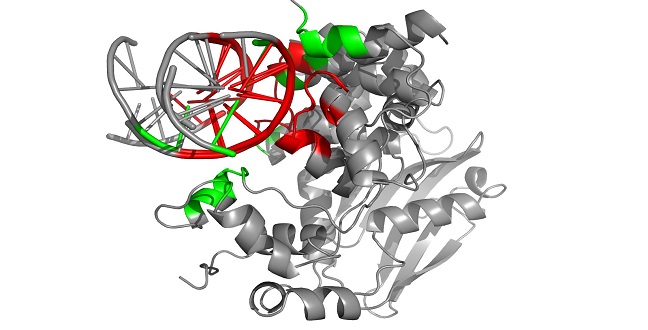
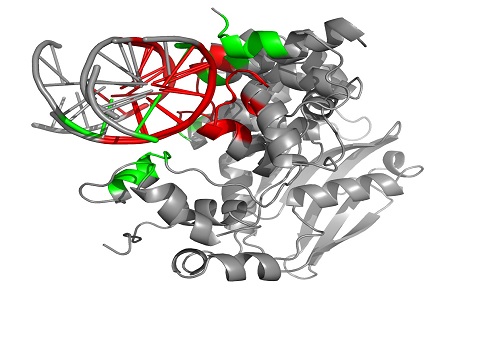
The PROSTA-inter program first finds the DNA binding interfaces (highlighted in green and red) of two molecular complexes (3OH9 and 2BZF) and then identifies the regions shared (highlighted in red) between the two interaction interfaces.
© 2015 KAUST
Understanding the form and function of molecular complexes has important implications for the development of new drugs. Researchers from King Abdullah University of Science and Technology (KAUST) have devised a method to uncover detailed information about molecular complexes while overcoming the practical challenges of existing methods1.
Biological molecules such as proteins, RNA and DNA perform their functions via interactions with other molecules, often forming connected “bundles” known as complexes. It is well-established that when these molecular complexes share a similar form, they are also likely to share a similar function.
Xin Gao, KAUST Assistant Professor of Computer Science, and colleagues from the University’s Computer, Electrical and Mathematical Science and Engineering Division have developed a method called PROSTA-inter. The method can align the regions of individual molecules that connect together (known as the interaction interfaces) of two molecular complexes according to their 3D conformations.
Programs that can structurally align individual molecules allow scientists to match those that share similar functions. However, existing technology cannot align differing molecular complexes. For example, in aligning a protein-protein complex with a protein-RNA complex, existing techniques would overlook at least one of the interacting partners.
“Although the global structures of interacting pairs may be very different, they could have similar interaction interfaces and therefore similar functions,” explained Gao. “Existing methods cannot handle the alignment of complexes in full. For example, a model of a protein-DNA complex may be flawed because the program does not acknowledge the fact that the two DNA chains take turns to interact with the protein.”
PROSTA-inter was developed to fill this gap in alignment software. The program searches for optimal structural matches between interaction interfaces and can analyze multiple chains and orientations of the molecules and their constituent parts.
“As you can imagine, the search space for all possible alignments is huge and difficult to explore,” Gao noted. “We overcame this challenge by demonstrating that the global optimal alignment can be captured by certain local alignment. We sampled local alignments to encode ‘fingerprints’ of molecular contact points before clustering them together to find near-optimal global alignments.”
The new method has already uncovered new details of structural similarities in two examples of functionally related protein-DNA complexes. Further analysis of a protein-protein complex and protein-RNA complex already known to scientists as a case of “protein-RNA mimicry” also revealed structural similarities between the two complexes.
The researchers believe that PROSTA-inter will enable further research into the functions and interactions of molecular complexes and could be a part of future drug discovery.
References
- Cui, X., Naveed, H., & Gao, X. Finding optimal interaction interface alignments between biological complexes. Bioinformatics 31, i133- i141 (2015). | article
You might also like

Bioengineering
Bio-inspired network structures for next-generation AI

Computer Science
Green quantum computing takes to the skies

Computer Science
Probing the internet’s hidden middleboxes

Bioscience
AI speeds up human embryo model research
Computer Science
Improving chip design on every level

Computer Science
Sweat-sniffing sensor could make workouts smarter

Computer Science
A blindfold approach improves machine learning privacy
Computer Science



