Bioscience
Helping flowing cells make a connection
A modern twist on a traditional biochemistry technique lets scientists study interactions that trap cells in circulation.
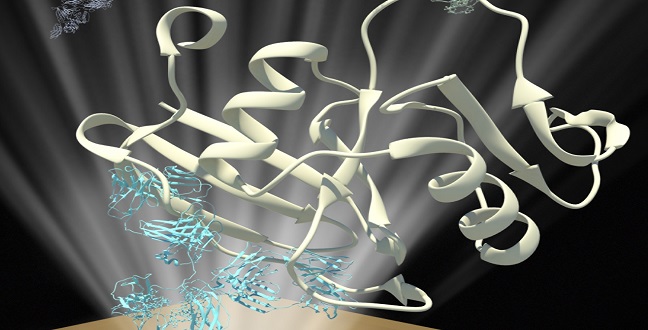
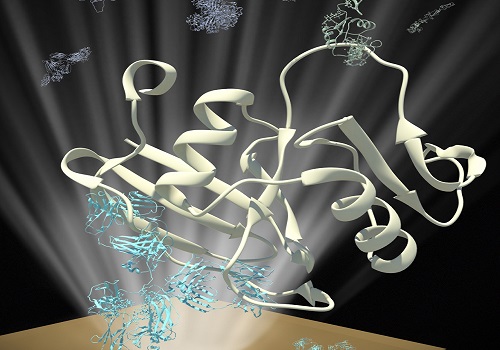
Schematic of an assay for measuring the interaction between E-selectin proteins (green) and their binding partners (white) that have been captured by antibodies (white) immobilized on a surface plasmon resonance chip.
© 2015 KAUST
Healthy blood stem cells and invasive tumor cells use similar mechanisms to exit the circulatory system and penetrate other tissues. An experimental strategy developed by Assistant Professor of Bioscience Jasmeen Merzaban and colleagues at KAUST allows for quantitative study of the earliest stages of this process.
“Cell migration is a multi-step process in which a number of adhesion molecules find their partners and mediate interaction of cells in the flow to the endothelial cells lining blood vessels,” explains Merzaban.
Her team is particularly interested in an endothelial protein known as E-selectin, but it has been difficult to characterize how this adhesion molecule captures circulating cells.
One established method for studying protein-protein interactions is immunoprecipitation, in which researchers use antibodies to purify a specific protein and capture other physically-associated binding partners in the process. However, immunoprecipitation is ineffective at detecting weak or fleeting interactions and offers limited insight into the rates at which molecules bind and separate.
Dina AbuSamra, a Ph.D. student in Merzaban’s group, tried to counter these deficiencies by developing an immunoprecipitation assay based on surface plasmon resonance (SPR), a technique that can measure molecular interactions in real-time.
The researchers first prepared SPR sensors with antibodies that efficiently capture known binding partners of E-selectin. They then exposed the sensors to flowing solutions of extracts from cells that express this protein followed by purified E-selectin. They were then able to accurately measure the speed and durability with which E-selectin binds to two of its target proteins.
Their results departed notably from prior predictions, which suggested that E-selectin binds and releases its targets quickly. However, these studies were conducted under relatively artificial conditions and focused on individual or ‘monomeric’ molecules of E-selectin, a protein that often pairs off to form ‘dimers’ (larger assemblies).
“Our results suggest that these ligands bound monomeric E-selectin transiently, with fast on- and off-rates, but bound dimeric E-selectin with remarkably slow on- and off-rates,” explains Merzaban. This indicates a process in which rapid but weak interactions with individual E-selectins initially capture circulating target cells and slow them sufficiently to form more durable connections with E-selectin dimers.
Their data also reveals how carbohydrate molecules on CD44 contribute to E-selectin binding. Merzaban’s team is now working to further dissect the molecular-scale details of this interaction mechanism to be able to manipulate it for clinical purposes.
“Our aim is to direct the delivery of cells, such as therapeutic stem cells, to sites where they are needed,” she says.
References
AbuSamra, D.B., Al-Kilani, A., Hamdan, S.M., Sakashita, K., Gadhoum, S.Z. & Merzhan, J.S. Quantitative characterization of E-selectin interaction with native CD44 and PSGL-1 using a real-time immunoprecipitation-based binding assay. Journal of Biological Chemistry jbc-M114 (2015).| article
You might also like

Bioscience
Robust workflow built for chemical genomic screening

Bioscience
Cell atlas offers clues to how childhood leukemia takes hold

Bioscience
Hidden flexibility in plant communication revealed

Bioscience
Harnessing the unintended epigenetic side effects of genome editing

Bioscience
Mica enables simpler, sharper, and deeper single-particle tracking

Bioengineering
Cancer’s hidden sugar code opens diagnostic opportunities

Bioscience
AI speeds up human embryo model research
Bioscience



