Plant Science
Stressing the genetics in salt-tolerant crops
Unravelling the genes behind salt tolerance in crop plants is made easier using the latest sequencing and phenotyping technologies.

The latest high-throughput genetic sequencing and phenotyping technologies will have a lasting, positive impact on plant science and crop sustainability research. KAUST researchers have conducted a timely review of these methodologies to highlight how they can help uncover the genes that underpin salt tolerance and other useful plant traits.
“Salt-tolerant crops promise greater food production using less freshwater—a win-win in the intensifying struggle for global food and water security,” says Mitchell Morton, who compiled the review with Mark Tester and colleagues at KAUST. “Recent technological advances make the development of crops with desirable traits more feasible than ever.”
Salt stress can have a huge impact on plant performance, ultimately affecting overall crop yields. An excess of salt can impede water uptake, reduce nutrient absorption and result in cellular imbalances in plant tissues. Plants have a systemic response to salt stress ranging from sensing and signaling to metabolic regulation. However, these responses differ widely within and between species, and so pinpointing associated genes and alleles is incredibly complex. Researchers must also disentangle other factors influencing genetic traits, such as local climate and different cultivation practices.
The KAUST team use state-of-the-art technologies to conduct their work in plant genetics research and sustainable agriculture. Genome-wide association studies, commonly used to scan genomes for genetic variants associated with specific traits, will help to determine the genes and mutations responsible for individual plant responses.

A Smarthouse at The Plant Accelerator (a high-throughput phenotyping system built under Prof. Mark Tester’s leadership) during an experiment on S. pimpinellifolium.
© 2019 Mitchell Morton
Another area of interest involves using unmanned aerial vehicles (UAVs) to scan fields and measure plant characteristics. Conducting such large-scale plant phenotyping manually is painstaking and provides limited data.

The M600 UAV, mounted with a hyperspectral camera, in mid-flight above a field experiment to measure the salt stress responses of 200 accessions of wild tomato S. pimpinellifolium.
© 2017 KAUST Halo Lab
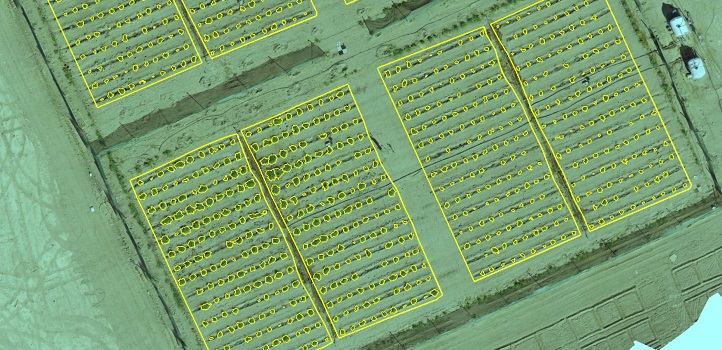
UAV-imagery-derived orthomosaic of a S. pimpinellifolium saline field experiment with automated delineation of individual plants and plots.
© 2019 KAUST HALO lab
“Using UAVs, we can collect meaningful data on thousands upon thousands of plants grown in the field, from biomass to growth rates, flowering time to yield,” says Morton. “We recently deployed UAV-based systems to phenotype nearly 200 accessions of the wild tomato Solanum pimpinellifolium in the field, using RGB, thermal and hyperspectral imaging. Our preliminary results look very promising.”
This increase in technology comes hand-in-hand with vast amounts of data. While there are various software and analytical tools for processing big data in plant science, none of them provide all the features that might be required.
“I would like to see more integrated solutions emerge. The easier they are to use, the more likely it is for these technologies to be adopted by people beyond academia,” says Morton.
References
-
Morton, M., Awlia, M., Al-Tamimi, N., Saade, S., Pailles, Y. , Negrão, S & Tester, M. Salt stress under the scalpel – dissecting the genetics of salt tolerance. The Plant Journal 97, 148-163 (2018).| article
You might also like

Plant Science
Coevolution of the microbiome with its host plant supports environmental adaptation

Bioscience
Hidden flexibility in plant communication revealed

Plant Science
Reference genomes for rice’s wild relatives may boost future crops
Bioscience
Digging into the world of plant-growth-promoting microbes

Environmental Science and Engineering
Hydrogen storage solution could lie in lakes

Bioscience
Unraveling modern bread wheat from the genes up

Bioscience
Why do some plants thrive in saline conditions?
Bioengineering




