Plant Science
Molecular immunity from microbes
Harnessing a bacterial system of defense can protect plants against viral pathogens.
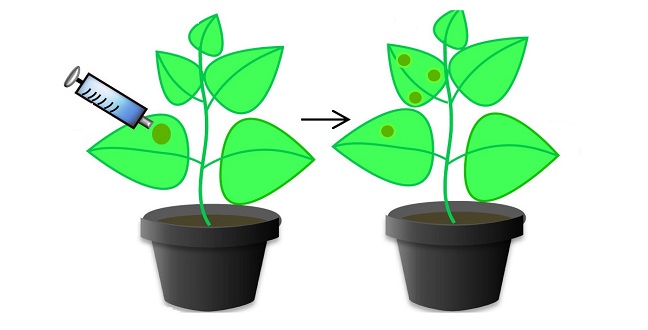
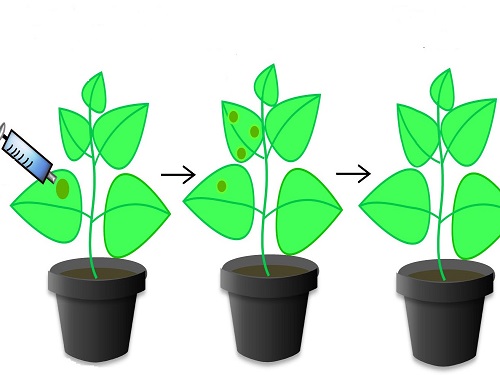
The tobacco plant Nicotiana benthamiana has been engineered to include the CRISPR/Cas9 system that provides resistance to tomato yellow leaf curl virus.
Modified and reproduced with permission from reference 1. © 2015 KAUST
A new molecular biology tool derived from a bacterial defense system has been used for the first time by KAUST researchers to demonstrate a novel way to protect plants against viral pathogens1.
Viruses are a major cause of crop disease, and infection by them results in enormous financial losses each year. They consist only of genetic material (DNA or RNA) surrounded by a protective coat. Lacking any cellular machinery of their own, they exploit the mechanisms of their host cells to replicate.
In bacteria, the CRISPR/Cas9 system provides immunity to DNA-based viruses by recognizing and disrupting their DNA. It is also rapidly gaining prominence as an effective new tool of modern molecular biology as a means to induce mutations and therefore novel characteristics in plant breeding.
The KAUST team, which was led by Magdy Mahfouz, introduced the Cas9 component — an enzyme capable of cleaving DNA — into tobacco plants. The team also engineered the plants to contain a short molecule known as a sgRNA, which is designed to complement part of the DNA sequence of the tomato yellow leaf curl virus (TYLCV). When the plants were infected with TYLCV, the sgRNA directed the Cas9 enzyme to attack the viral DNA, preventing it from replicating and significantly reducing disease symptoms.
Protection against TYLCV, a significant scourge of tomato crops, was only the first step. The researchers next engineered plants with multiple identical sgRNAs. This process generated even stronger resistance to TYLCV, an effect they termed multiplexing. They finally inserted a new sgRNA matching the geminiviruses, a whole family of viruses, thereby providing simultaneous protection against multiple diseases.
The research demonstrates that this simple microbial system can provide crop plants with a powerful defense against multiple invading viruses. The system can even be fine-tuned to defend against newly emerging viral strains simply by tweaking the sgRNA used.
Mahfouz notes that the work also provides insights into the molecular mechanisms of natural virus resistance in plants.
Identifying the missing pieces in the puzzle will “help us to generate plants that are resistant to viruses while remaining free of foreign DNA,” he said. “Such engineered plants would avoid the regulatory hurdles surrounding genetic modification and be more acceptable to a GM-averse public.”
The next step for the KAUST team is to modify the system to work on RNA-based as well as DNA-based viruses. Mahfouz is confident that this more difficult challenge is also surmountable. “It is only a matter of time,” he noted.
References
- Ali, Z., Abulfaraj, A., Idris, A., Ali, S., Tashkandi, M. & Mahfouz, M. CRISPR/Cas9-mediated viral interference in plants. Genome Biology 16, 238 (2015).| article
You might also like

Plant Science
Coevolution of the microbiome with its host plant supports environmental adaptation

Bioscience
Hidden flexibility in plant communication revealed

Plant Science
Reference genomes for rice’s wild relatives may boost future crops
Bioscience
Digging into the world of plant-growth-promoting microbes

Environmental Science and Engineering
Hydrogen storage solution could lie in lakes

Bioscience
Unraveling modern bread wheat from the genes up

Bioscience
Why do some plants thrive in saline conditions?
Bioengineering



