Marine Science
Tweaking the transcriptome to tackle stress
Stressed dinoflagellates rewrite their genes during transcription.
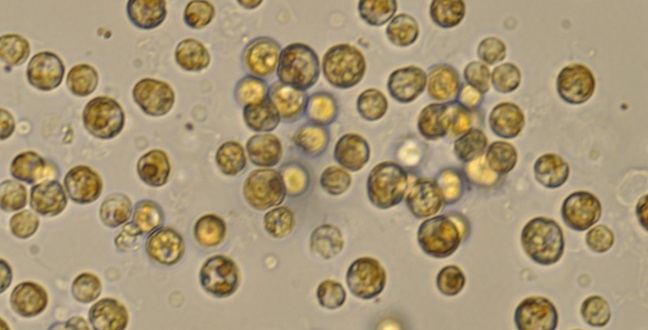
Symbiodinium cells viewed through a light microscope.
© 2017 Fabia Simona
Single-celled plankton known as dinoflagellates are shown to cope with stress using an unexpected strategy of editing their RNA rather than changing gene expression levels1.
The finding by KAUST researchers began when a team led by Associate Professor Christian Voolstra and Assistant Professor Manuel Aranda compared RNA transcripts from two strains of dinoflagellates thought to belong to the same species in the genus Symbiodinium. The transcripts had more differences than expected, indicating a more distant relationship. However, the team speculated that the RNA transcripts might instead have been edited, producing different information than that encoded in the cell’s DNA.
RNA editing had previously been observed in the mitochondria and plastids of dinoflagellates, but not in genes encoded in the nucleus. Earlier studies by Voolstra and Aranda had shown that gene expression changes very little in dinoflagellates under stressful conditions. The researchers wondered: “What if they do it completely differently? What if they just edit transcripts the way they need instead of changing expression?’” recalled Aranda.
To test their hypothesis, the team analyzed transcriptomes from Symbiodinium cultures grown in normal conditions and stressed by cold, heat or darkness. A conservative estimate uncovered 3,300 RNA edits. “This expands the encoding capacity beyond what’s in the genome,” said Aranda, effectively giving the cell a fuzzy genome. “Instead of having just one version of a protein, they can produce multiple different versions by changing the message on a different level.”
The team then turned its attention to 229 genes edited in all four growth conditions. The RNA of half of these genes was edited differently in at least one of the stressed cultures, and many of the genes had changes in RNA editing in response to several stress factors. “We’ve shown that short-term stress can be dealt with by RNA editing. But we don’t yet know how,” said the study’s lead author, Dr. Yi Jin Liew.
Understanding the mechanism behind RNA editing and how this machinery is regulated remains an exciting challenge for future research.
These findings also raise intriguing questions about the evolutionary and ecological implications of this process. RNA editing may offer dinoflagellates an efficient mechanism for rapid evolutionary experimentation.
The ecological implications may be broader still. Many Symbiodinium species are symbiotic with corals, and the coral’s stress tolerance partly depends on the identity of its Symbiodinium partner. “It would be very interesting to check whether coral-symbiont combinations that tolerate high temperatures undergo more or less RNA editing,” said Voolstra.
References
-
Liew, Y. J., Li, Y., Baumgarten, S., Voolstra, C. & Aranda, M. Condition-specific RNA editing in the coral symbiont Symbiodinium microadriaticum. PLoS Genetics 13, e1006619 (2017).| article
You might also like

Marine Science
Potential gains from replenishing reef fish stocks revealed

Marine Science
A place to trial hope for global reef restoration

Marine Science
Reef-building coral shows signs of enhanced heat tolerance

Marine Science
Plastic-munching bacteria found across the seven seas

Marine Science
AI reveals the universal beauty of coral reef growth

Marine Science
Tiny crabs glow to stay hidden

Marine Science
Mass fish deaths linked to extreme marine heatwave in Red Sea

Marine Science



